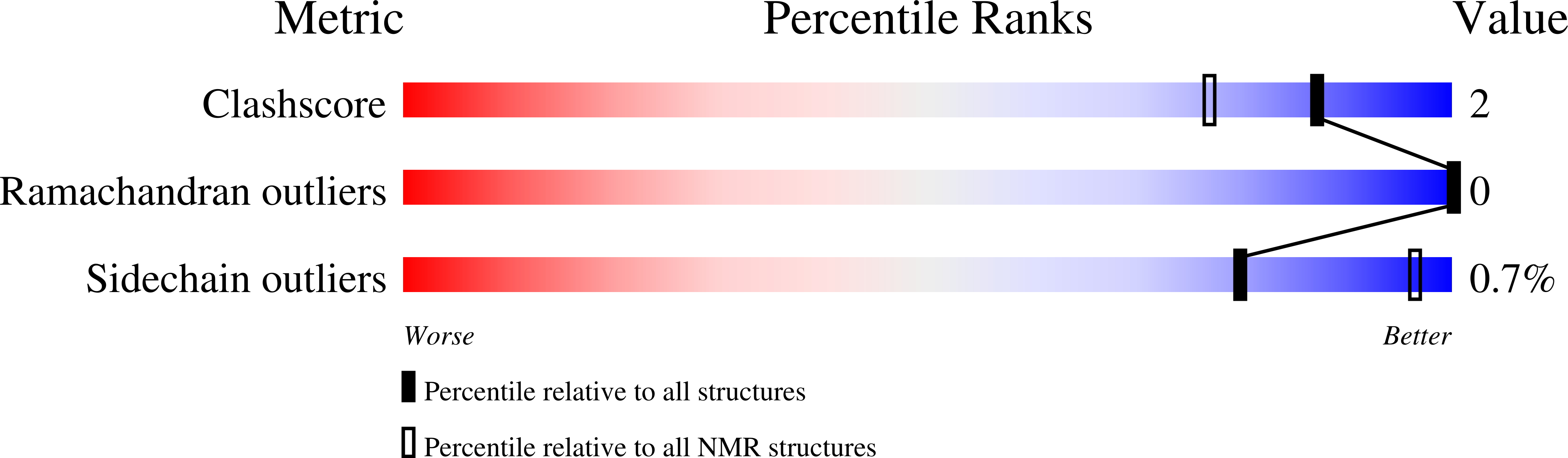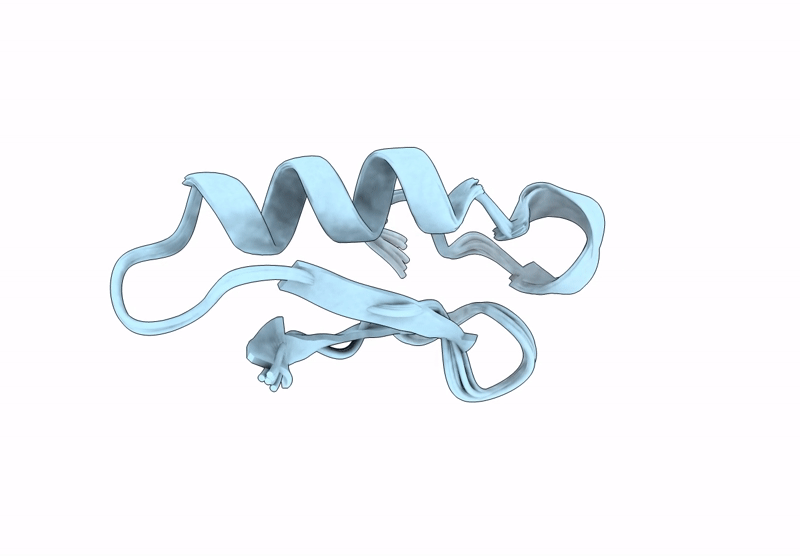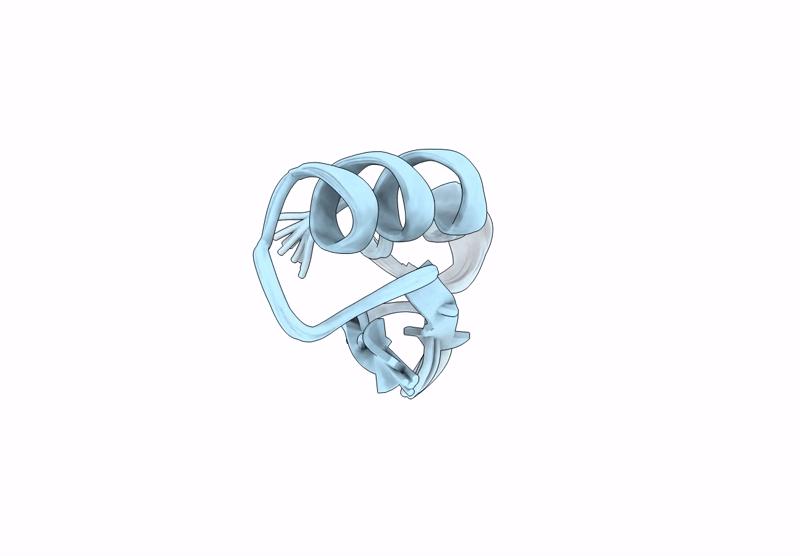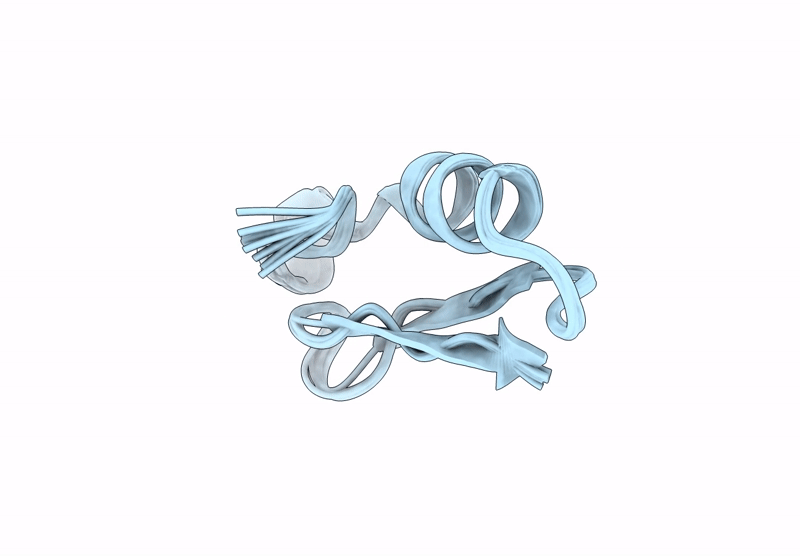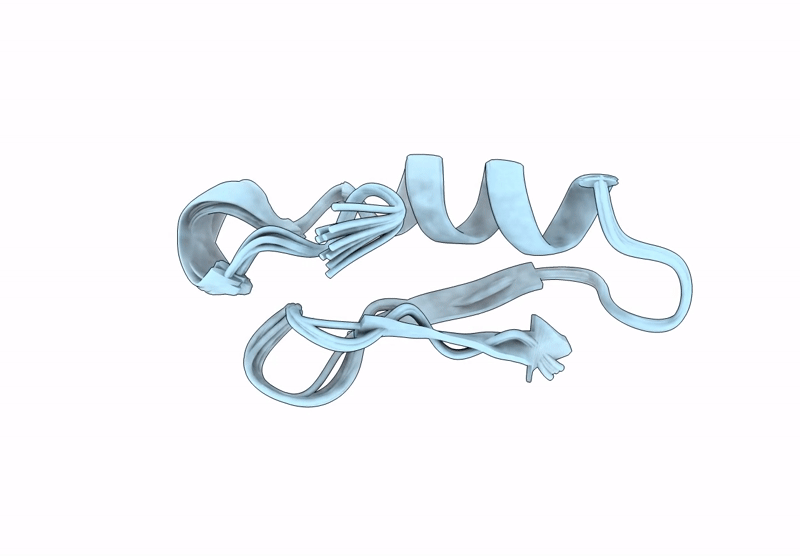
Deposition Date
2024-10-24
Release Date
2025-01-08
Last Version Date
2025-03-19
Entry Detail
PDB ID:
9E3Z
Keywords:
Title:
Backbone Modification in the Fungal Defensin Plectasin: Calpha-methyl-residues in the helix, D- and Calpha-methyl-residues in the turns
Biological Source:
Source Organism:
Pseudoplectania nigrella (Taxon ID: 96584)
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
10
Selection Criteria:
structures with the lowest energy