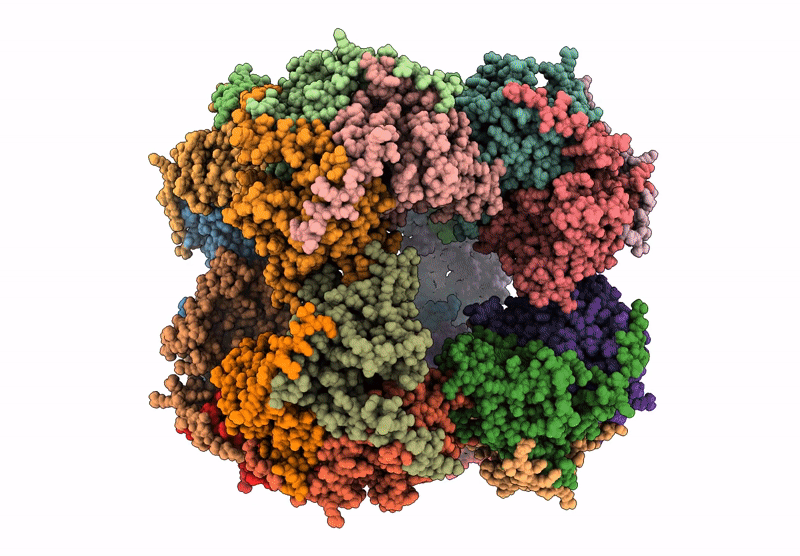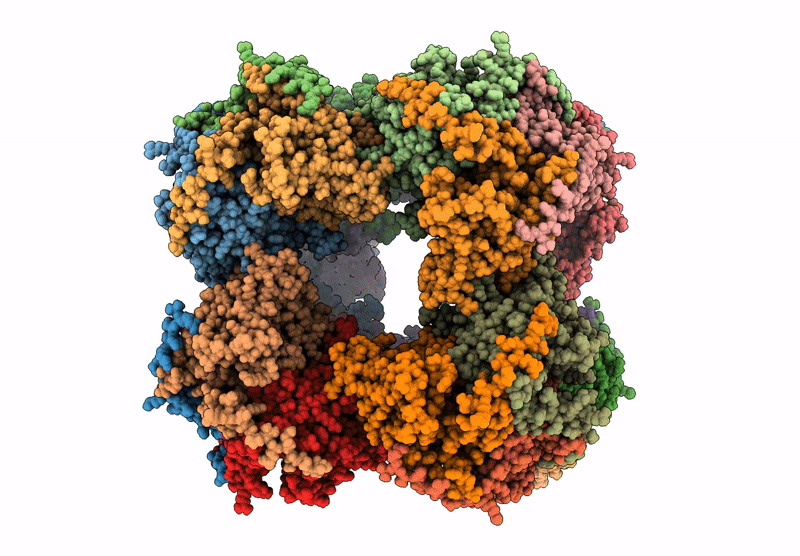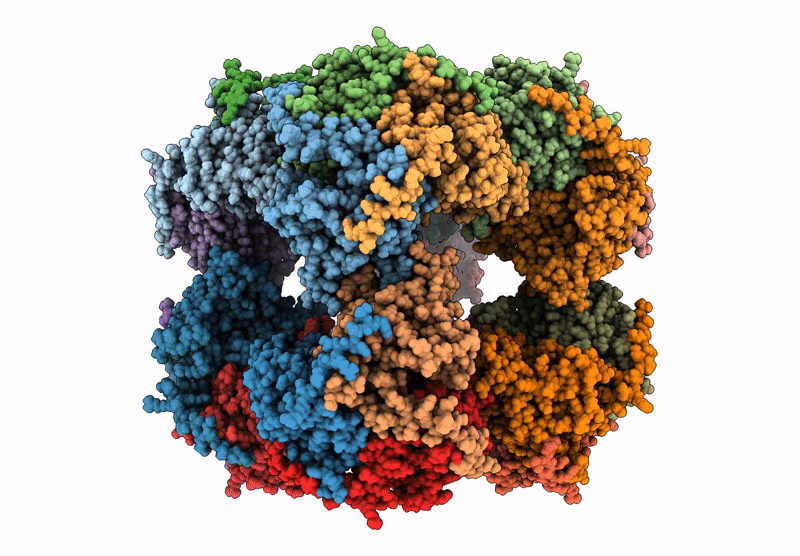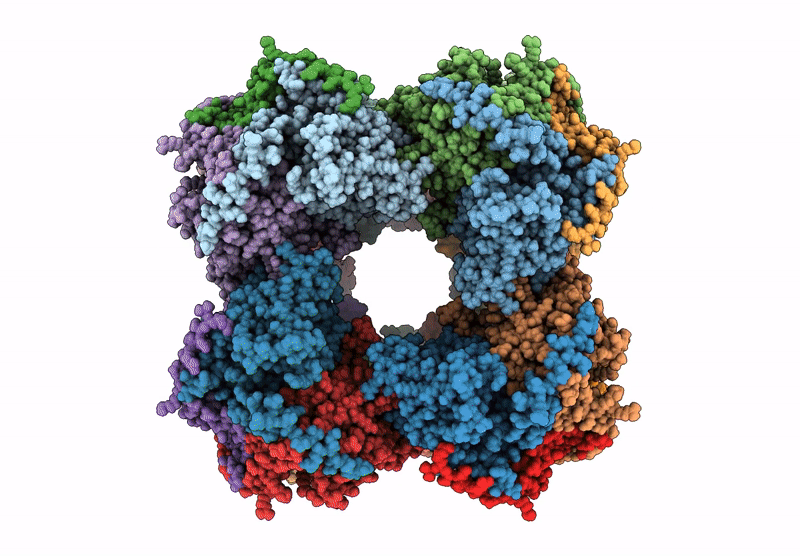
Abstact
Protein purification is essential in protein biochemistry, structural biology, and protein design, enabling the determination of protein structures, the study of biological mechanisms, and the characterization of both natural and de novo designed proteins. However, standard purification strategies often encounter challenges, such as unintended co-purification of contaminants alongside the target protein. This issue is particularly problematic for self-assembling protein nanomaterials, where unexpected geometries may reflect novel assembly states, cross-contamination, or native proteins originating from the expression host. Here, we used an automated structure-to-sequence pipeline to first identify an unknown co-purifying protein found in several purified designed protein samples. By integrating cryo-electron microscopy (Cryo-EM), ModelAngelo's sequence-agnostic model-building, and Protein BLAST, we identified the contaminant as dihydrolipoamide succinyltransferase (DLST). This identification was validated through comparisons with DLST structures in the Protein Data Bank, AlphaFold 3 predictions based on the DLST sequence from our E. coli expression vector, and traditional biochemical methods. The identification informed subsequent modifications to our purification protocol, which successfully excluded DLST from future preparations. To explore the potential broader utility of this approach, we benchmarked four computational methods for DLST identification across varying resolution ranges. This study demonstrates the successful application of a structure-to-sequence protein identification workflow, integrating Cryo-EM, ModelAngelo, Protein BLAST, and AlphaFold 3 predictions, to identify and ultimately help guide the removal of DLST from sample purification efforts. It highlights the potential of combining Cryo-EM with AI-driven tools for accurate protein identification and addressing purification challenges across diverse contexts in protein science.