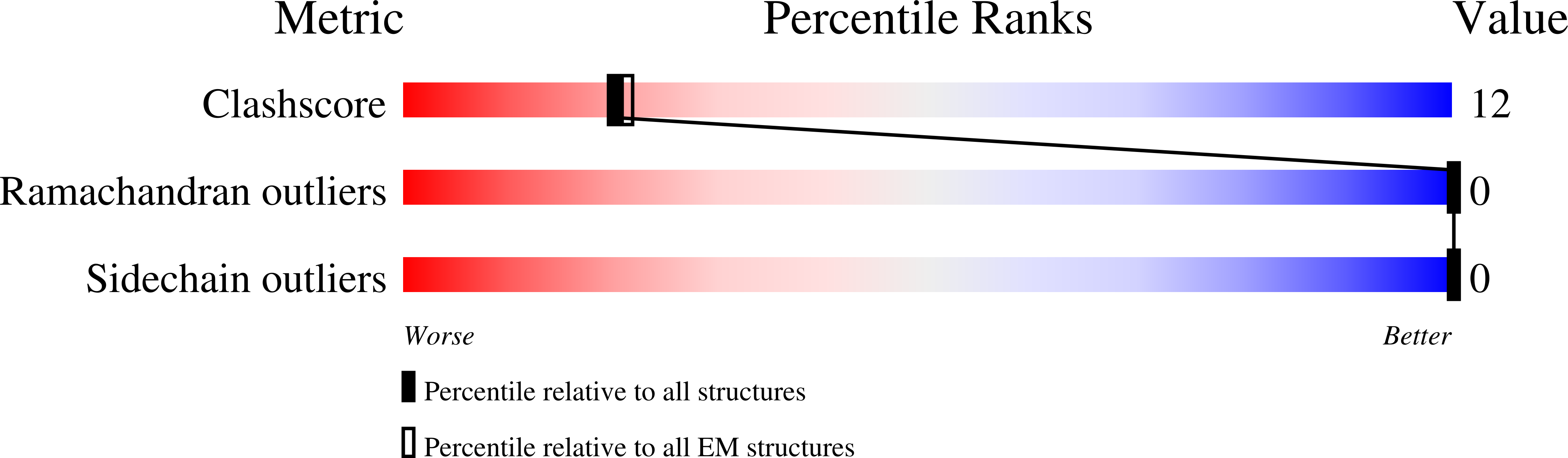
Deposition Date
2024-08-23
Release Date
2025-02-19
Last Version Date
2025-02-19
Entry Detail
PDB ID:
9DBN
Keywords:
Title:
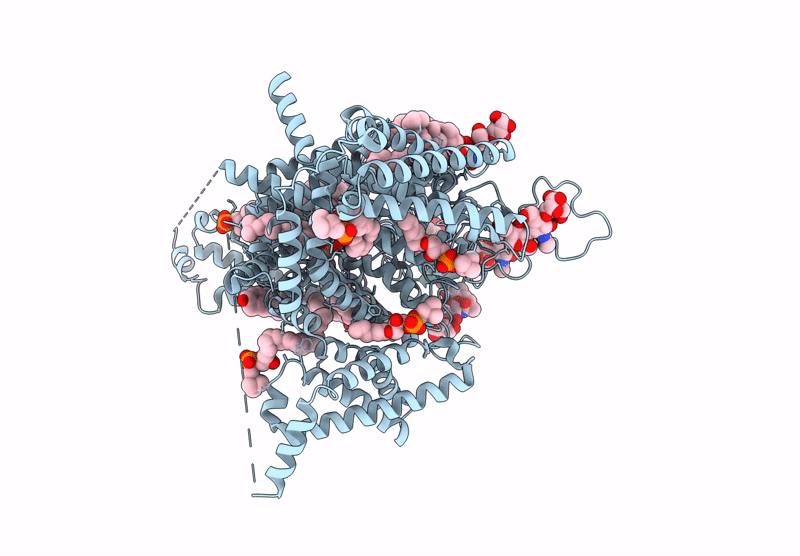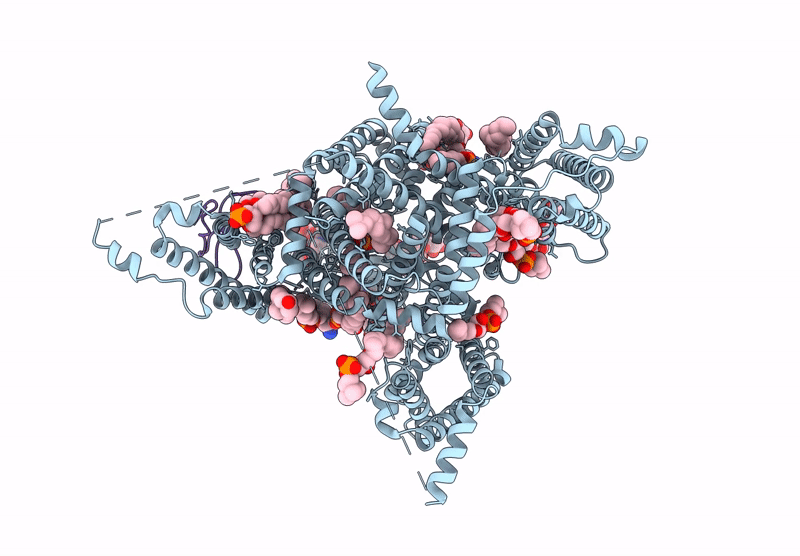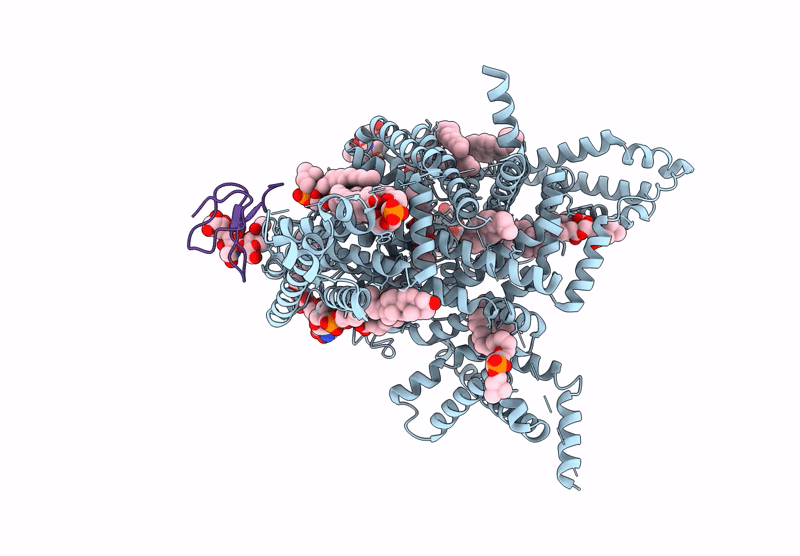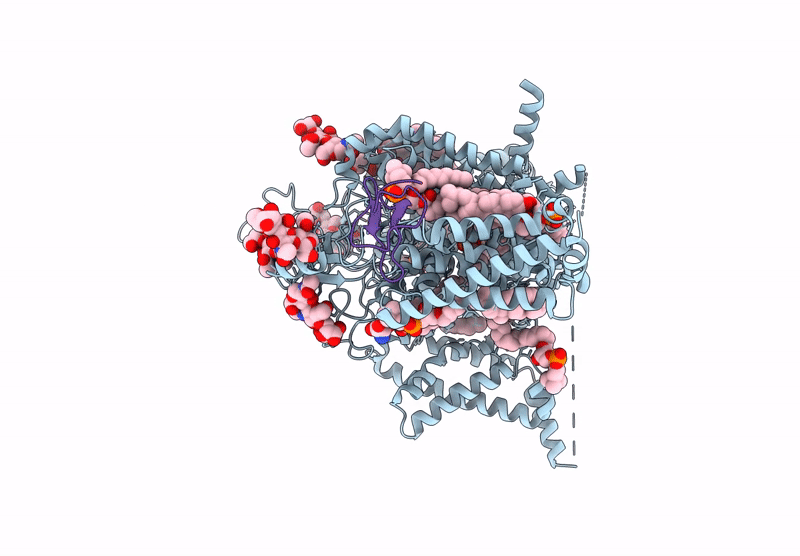
Tarantula venom peptide Protoxin-I bound to full-length human voltage-gated sodium channel 1.8 (NaV1.8)
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Thrixopelma pruriens (Taxon ID: 213387)
Thrixopelma pruriens (Taxon ID: 213387)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.76 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE