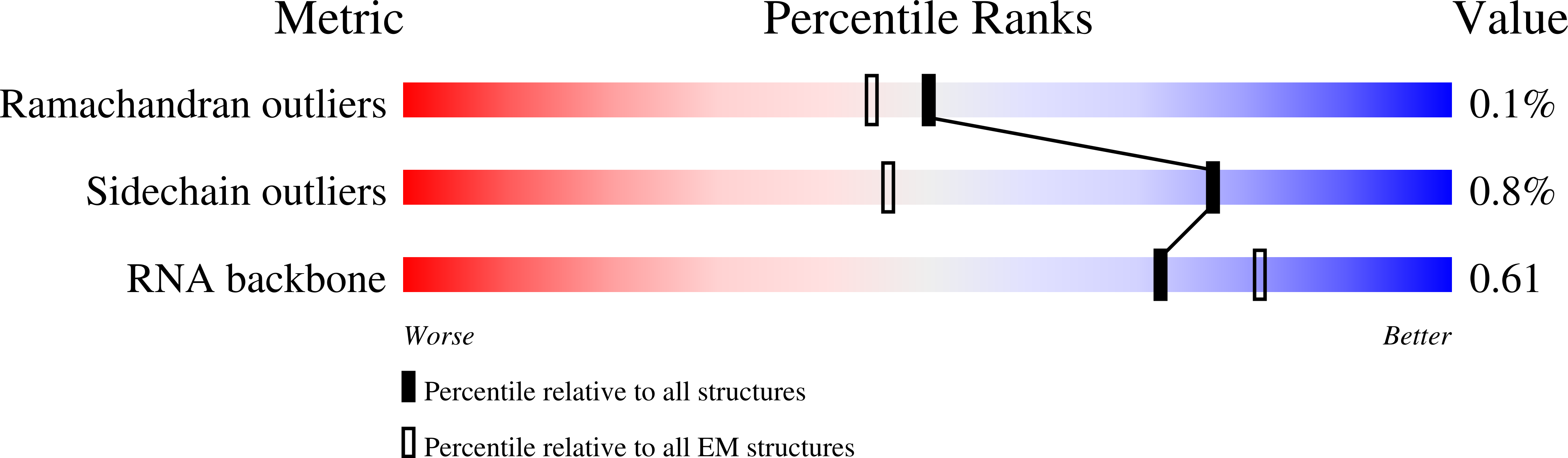
Deposition Date
2024-08-19
Release Date
2025-01-22
Last Version Date
2025-03-19
Entry Detail
PDB ID:
9D89
Keywords:
Title:
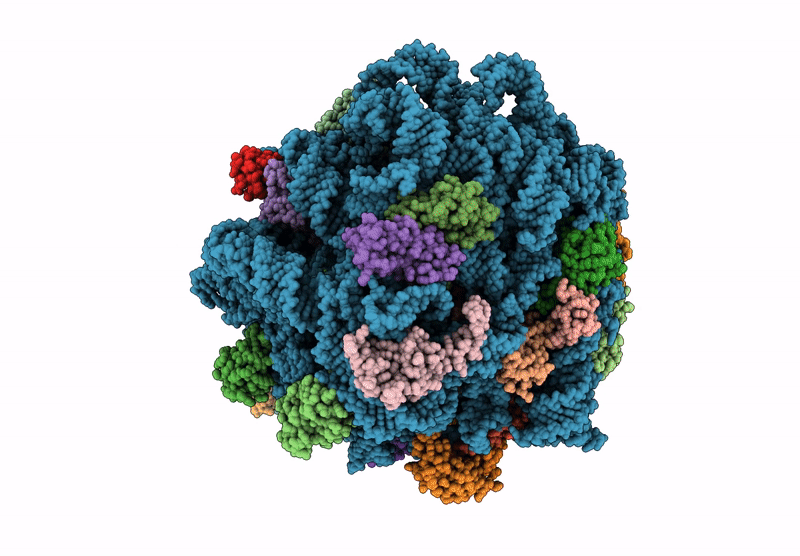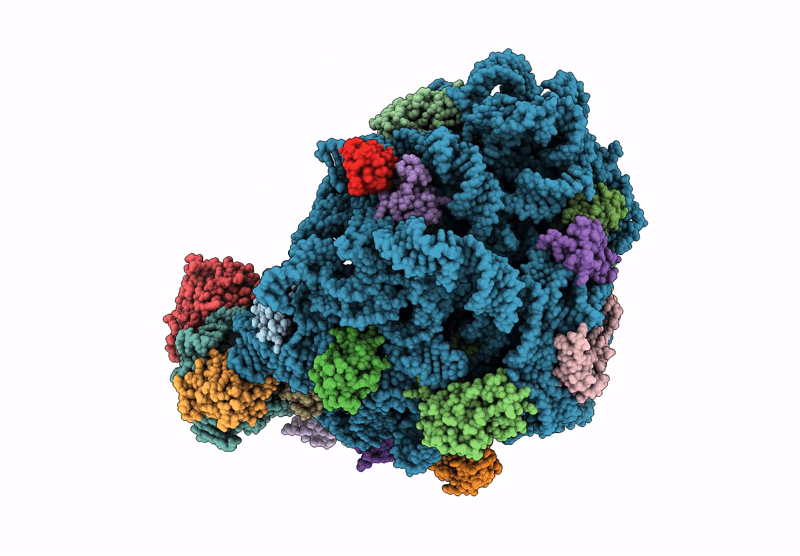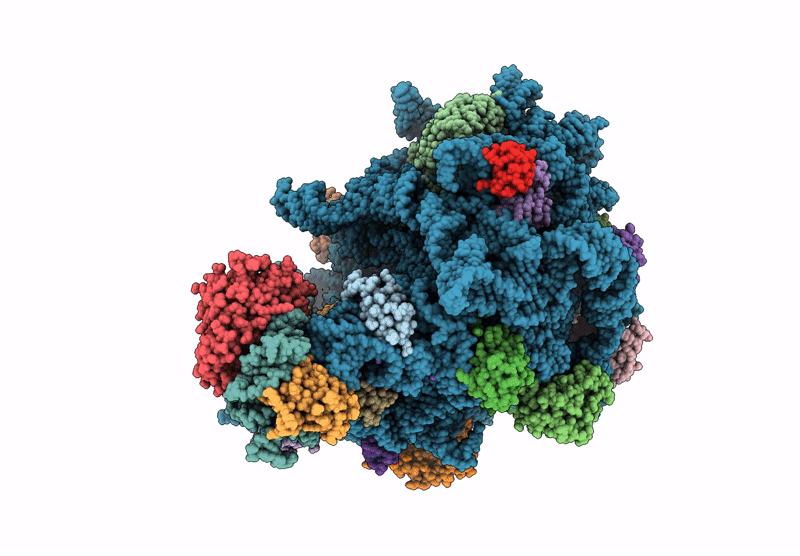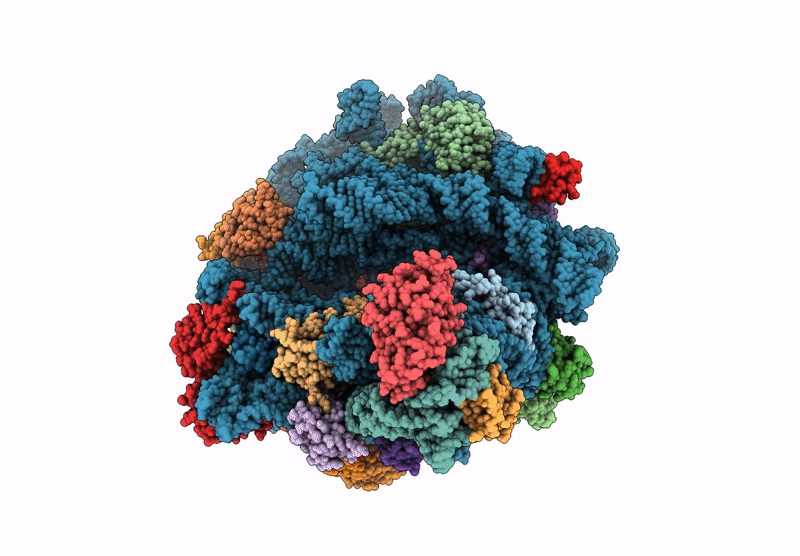
E. coli 50S ribosomal subunit in complex with PrAMP rumicidin-2 (focused refinement)
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Beatragus hunteri (Taxon ID: 59527)
Beatragus hunteri (Taxon ID: 59527)
Method Details:
Experimental Method:
Resolution:
1.95 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE