Deposition Date
2024-08-07
Release Date
2025-08-27
Last Version Date
2025-08-27
Entry Detail
PDB ID:
9D0H
Keywords:
Title:
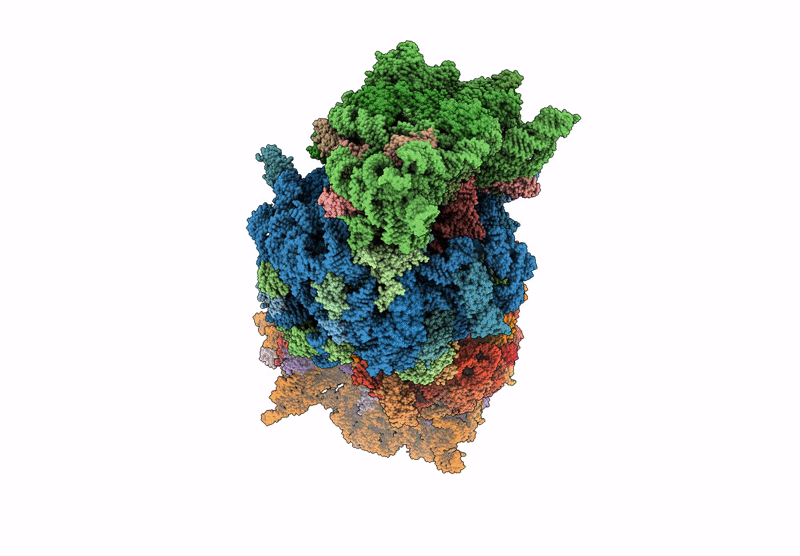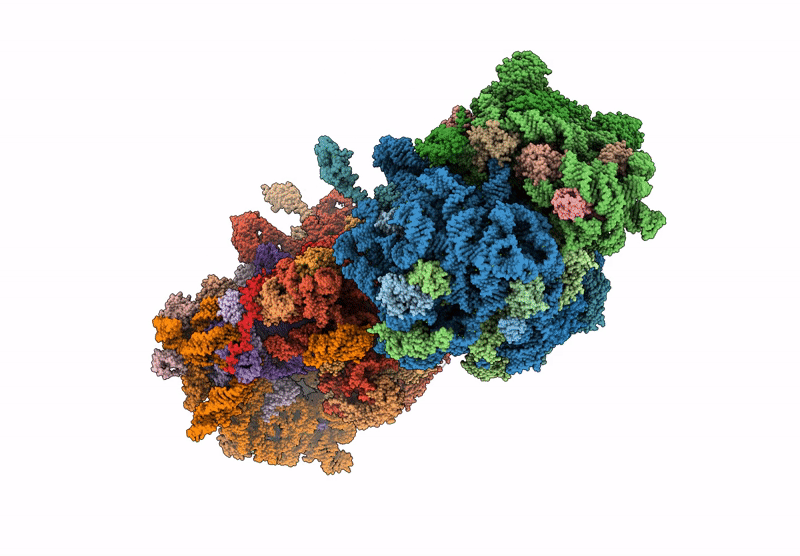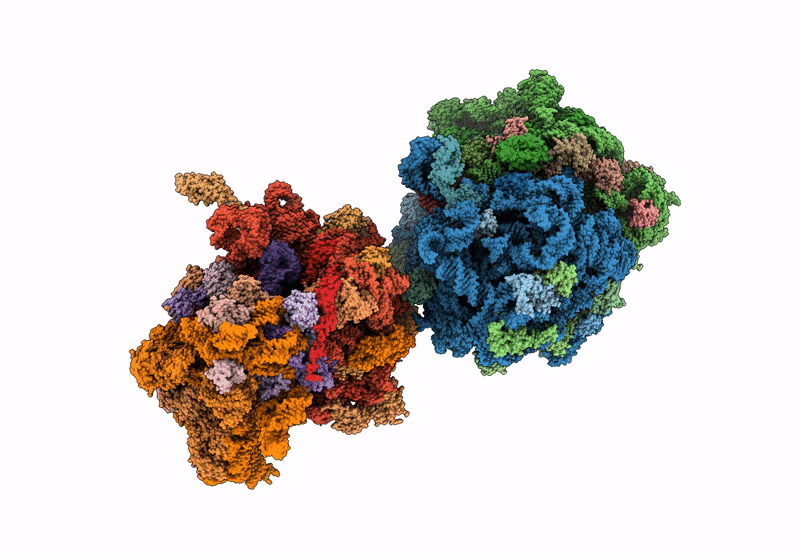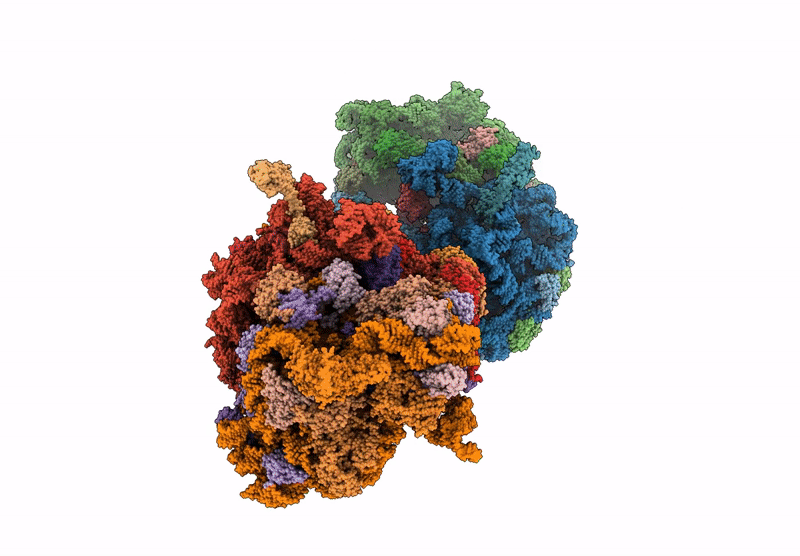
Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with C-cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Escherichia phage T4 (Taxon ID: 2681598)
Thermus thermophilus HB8 (Taxon ID: 300852)
Escherichia phage T4 (Taxon ID: 2681598)
Thermus thermophilus HB8 (Taxon ID: 300852)
Expression System(s):
Method Details:
Experimental Method:
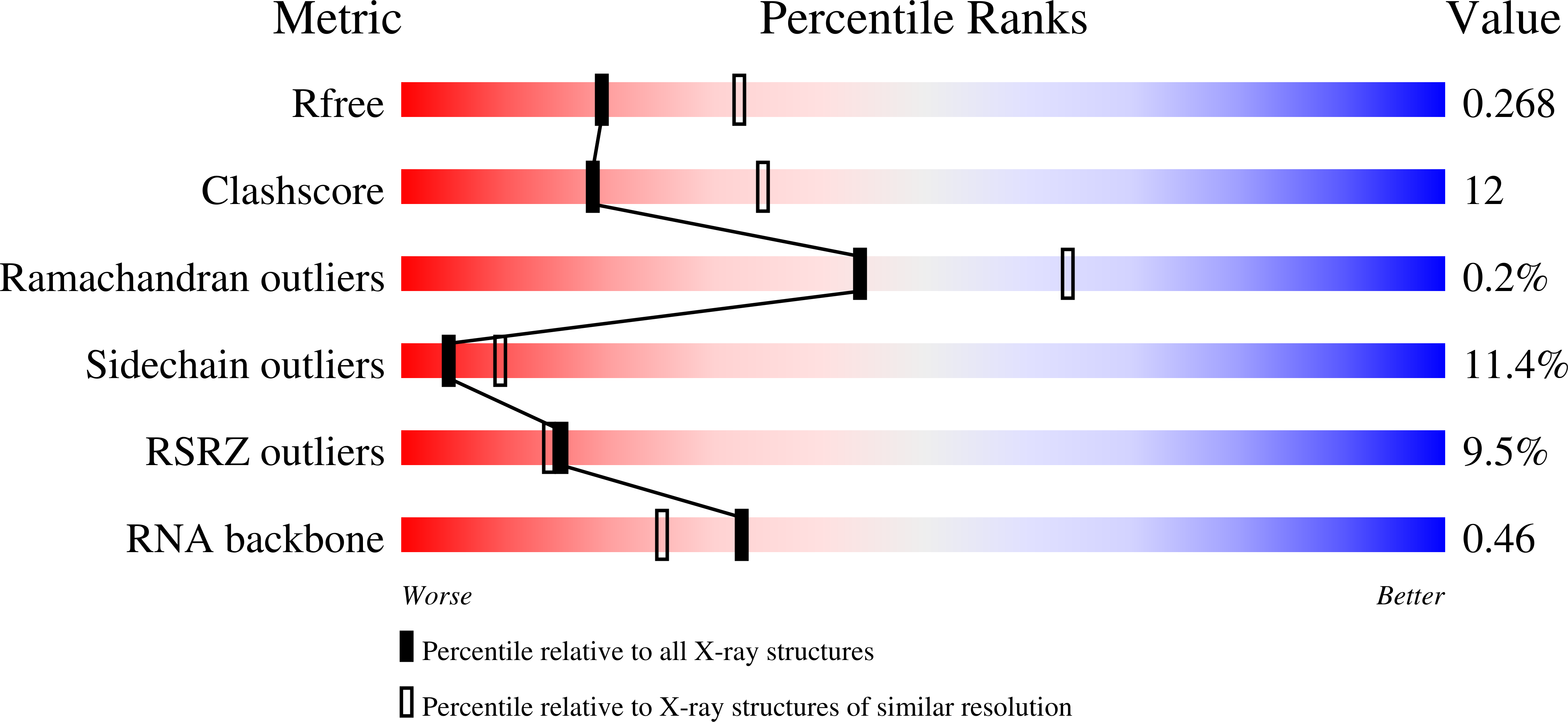
Resolution:
2.50 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21