Deposition Date
2024-07-24
Release Date
2024-12-11
Last Version Date
2025-01-01
Entry Detail
PDB ID:
9CSW
Keywords:
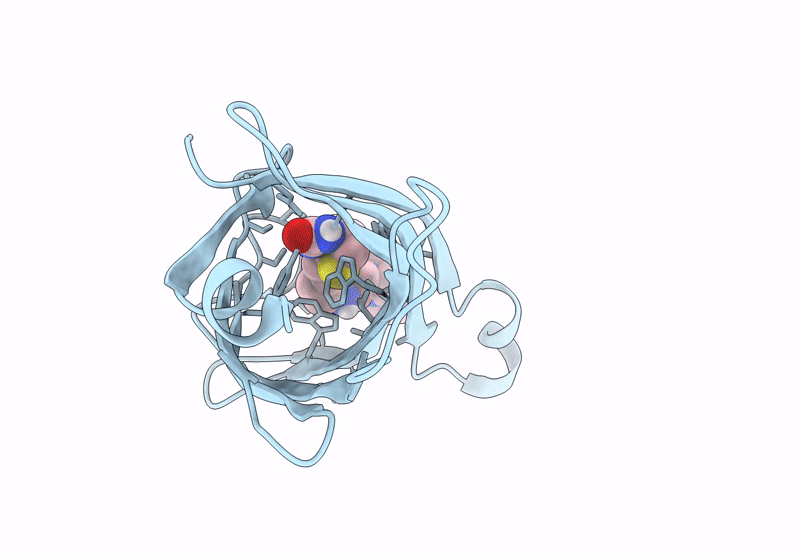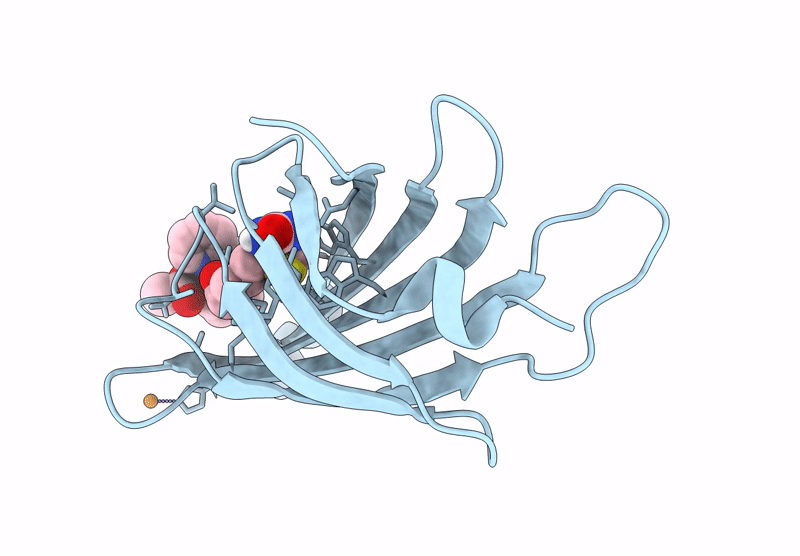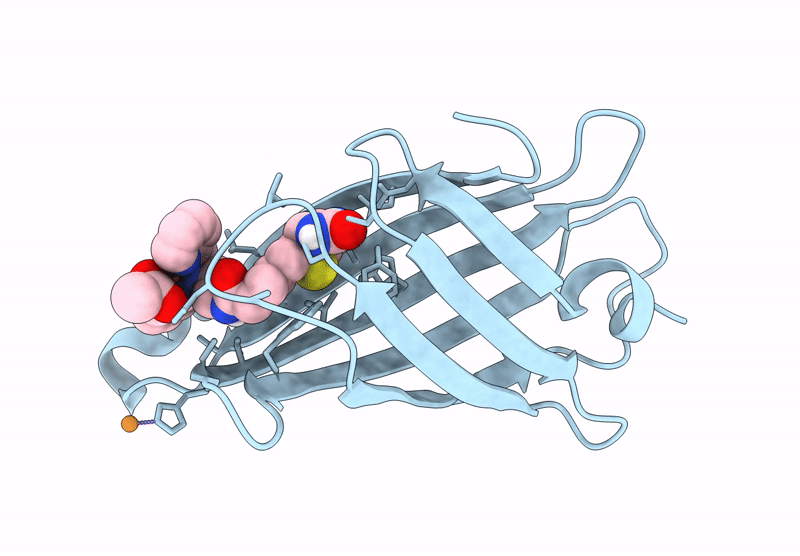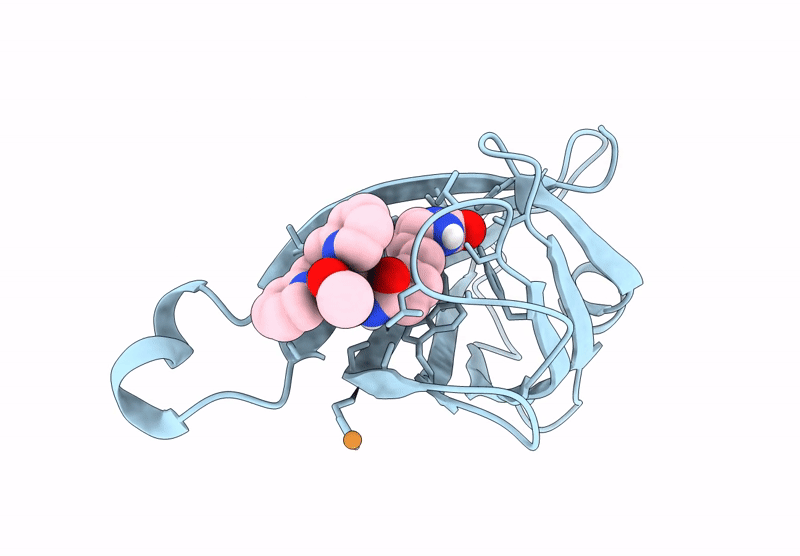
Title:
Streptavidin-E101Q-S112A-K121Y bound to Cu(II)-biotin-ethyl-dipicolylamine cofactor
Biological Source:
Source Organism(s):
Streptomyces avidinii (Taxon ID: 1895)
Expression System(s):
Method Details:
Experimental Method:
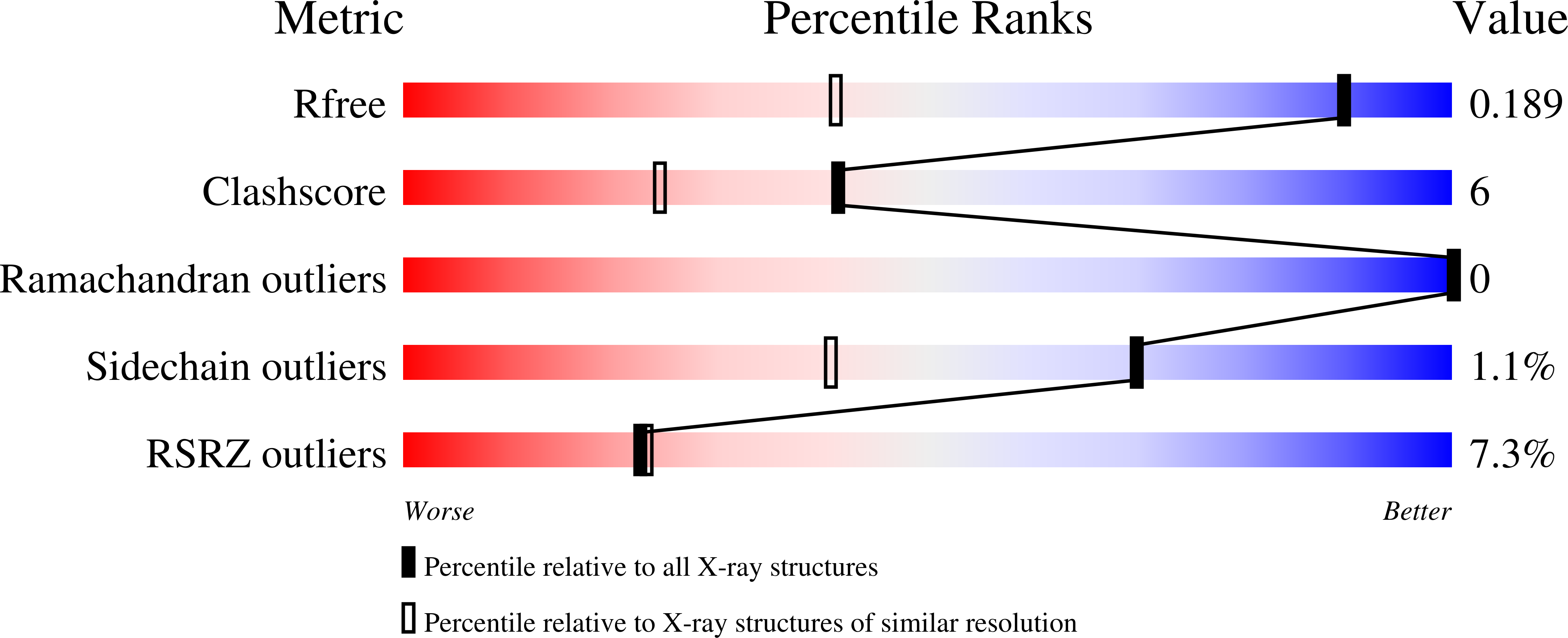
Resolution:
1.30 Å
R-Value Free:
0.18
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
I 41 2 2