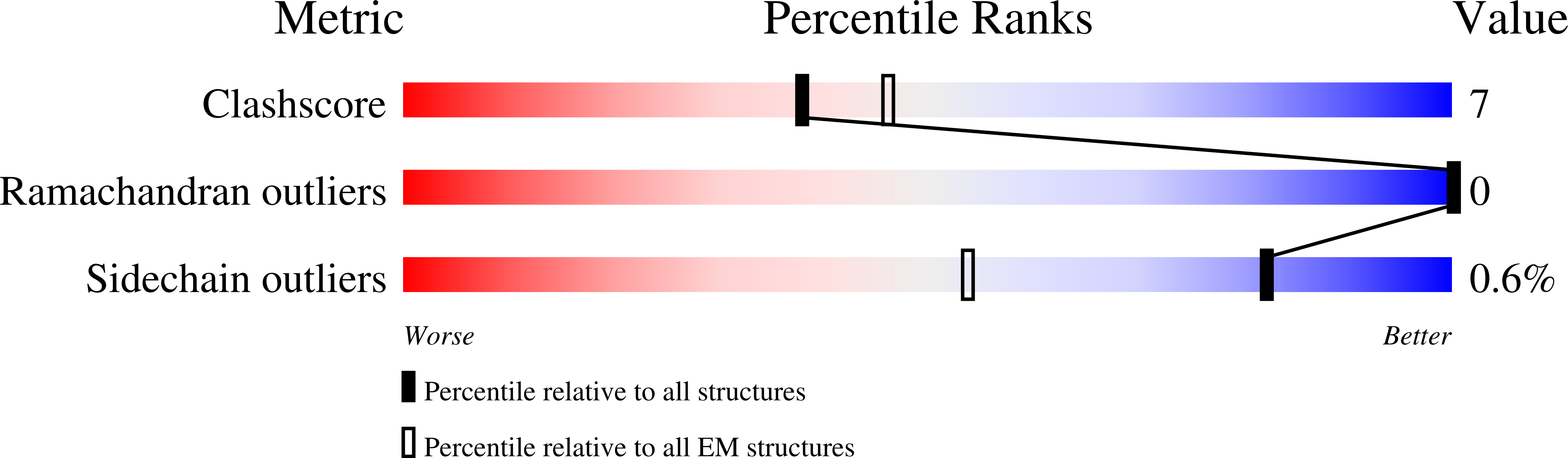
Deposition Date
2024-05-27
Release Date
2024-10-16
Last Version Date
2024-11-20
Entry Detail
PDB ID:
9C0S
Keywords:
Title:
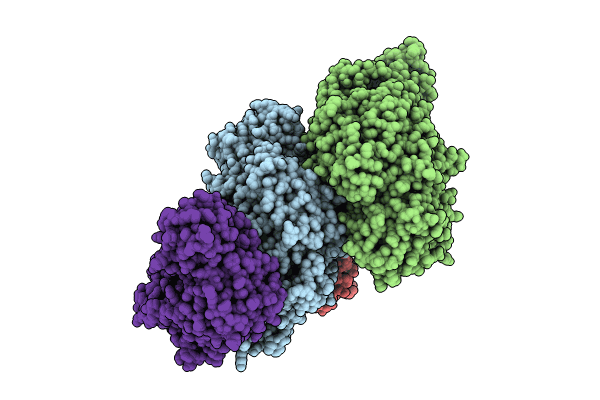
Carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) pentamer from Methanosarcina thermophila
Biological Source:
Source Organism(s):
Methanosarcina thermophila (Taxon ID: 2210)
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE