Deposition Date
2024-04-24
Release Date
2024-07-24
Last Version Date
2024-11-13
Entry Detail
PDB ID:
9BIS
Keywords:
Title:
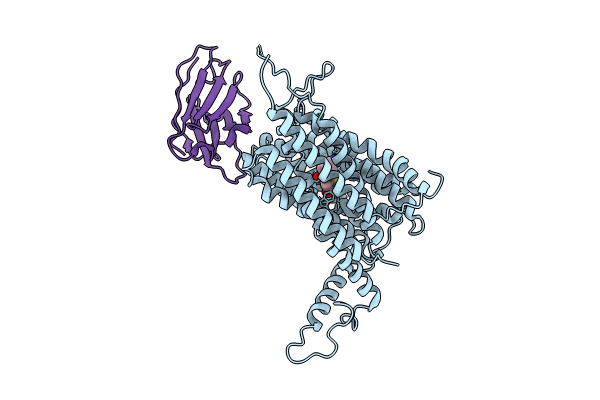
Cryo-EM structure of the mammalian peptide transporter PepT2 bound to amoxicillin
Biological Source:
Source Organism(s):
Rattus norvegicus (Taxon ID: 10116)
Lama glama (Taxon ID: 9844)
Lama glama (Taxon ID: 9844)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


