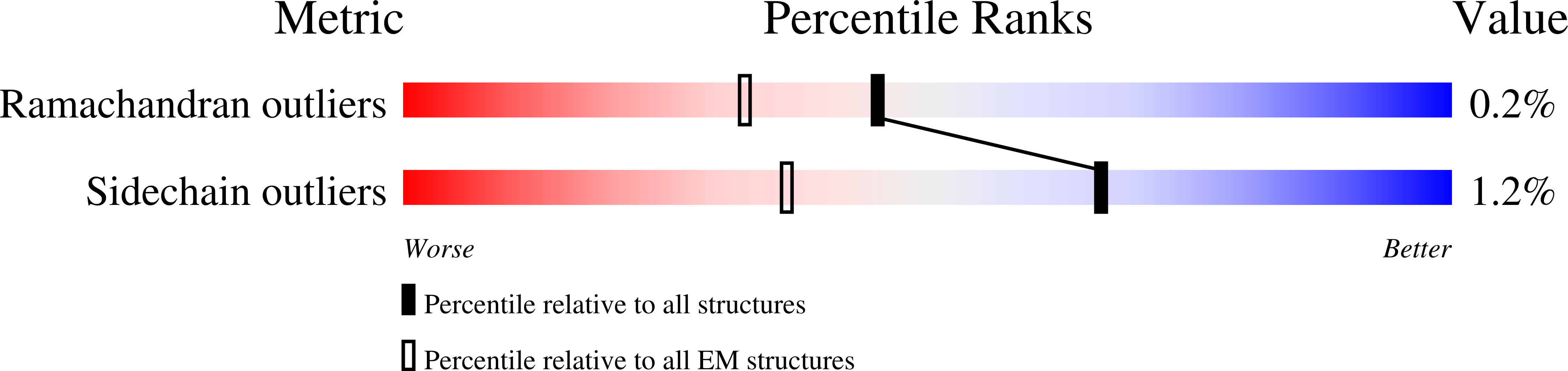
Deposition Date
2024-04-19
Release Date
2024-09-04
Last Version Date
2024-10-16
Entry Detail
PDB ID:
9BGM
Keywords:
Title:
Pseudomonas phage DEV neck and tail (portal, head-to-tail and tail tube proteins)
Biological Source:
Source Organism(s):
Pseudomonas phage vB_PaeP_DEV (Taxon ID: 2034344)
Method Details:
Experimental Method:
Resolution:
3.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE