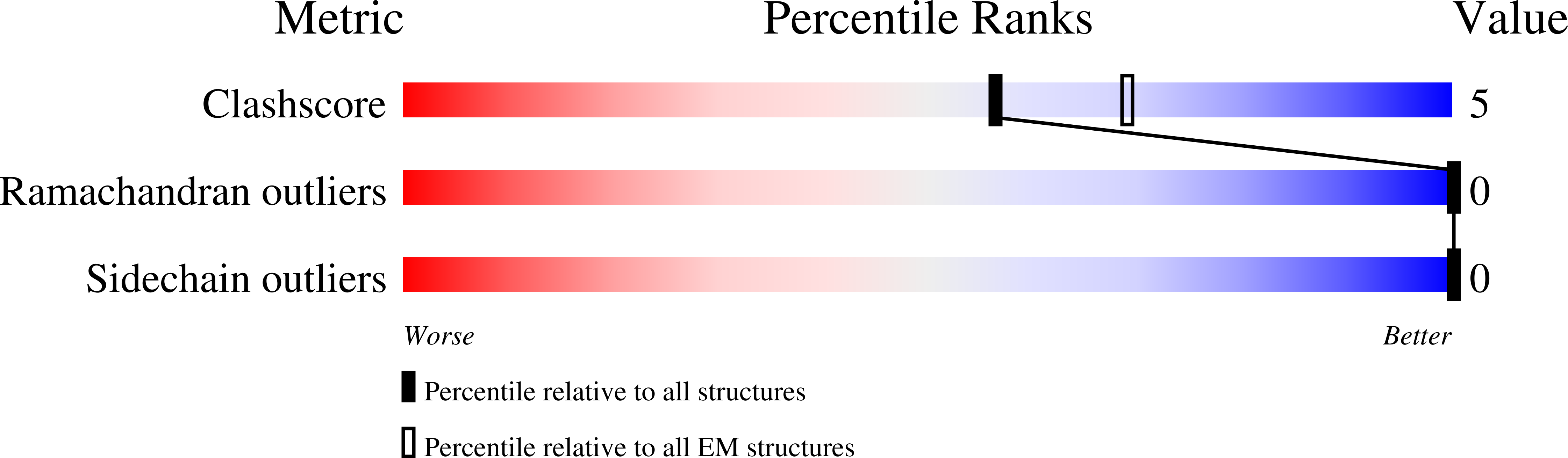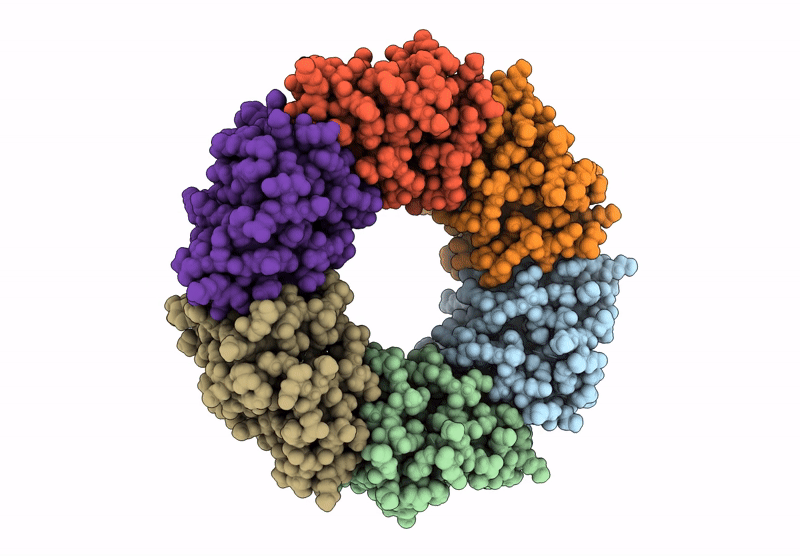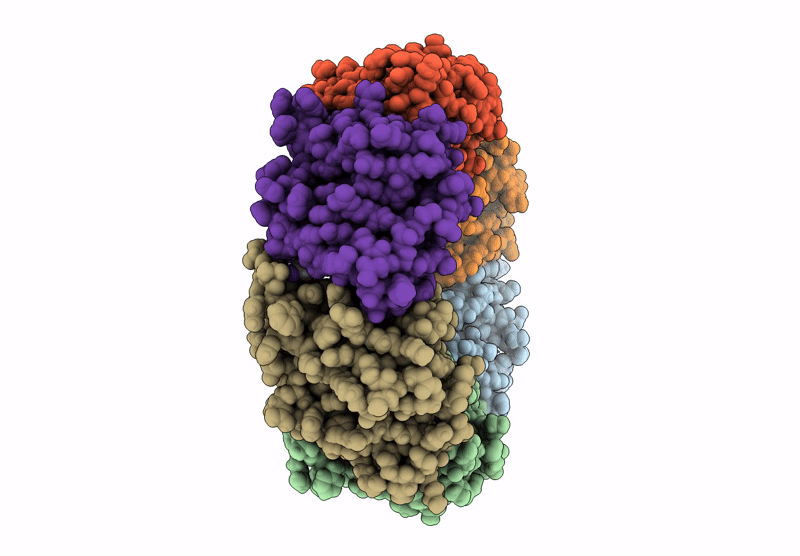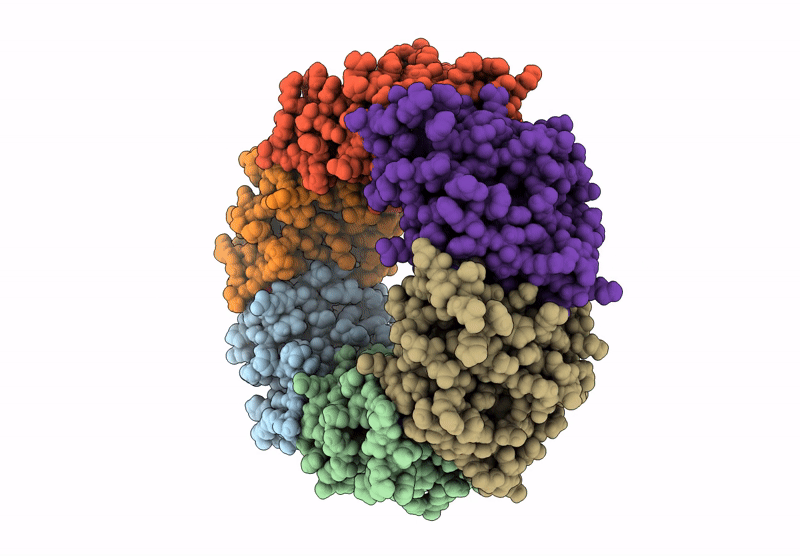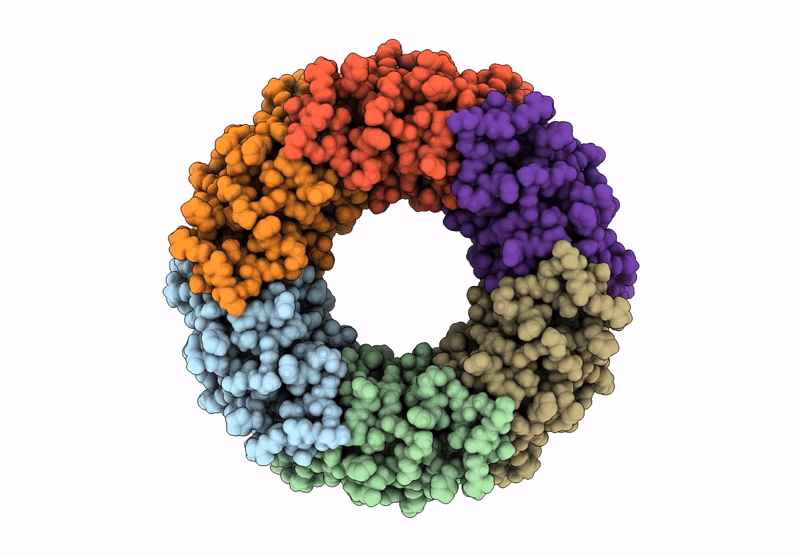
Deposition Date
2024-04-15
Release Date
2025-01-22
Last Version Date
2025-05-21
Entry Detail
PDB ID:
9BE8
Keywords:
Title:
Alkalihalobacillus halodurans (Aha) trp RNA binding attenuation protein (TRAP) mutant T49A/T52A dTRAP with Trp
Biological Source:
Source Organism(s):
Halalkalibacterium halodurans (Taxon ID: 86665)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.14 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE