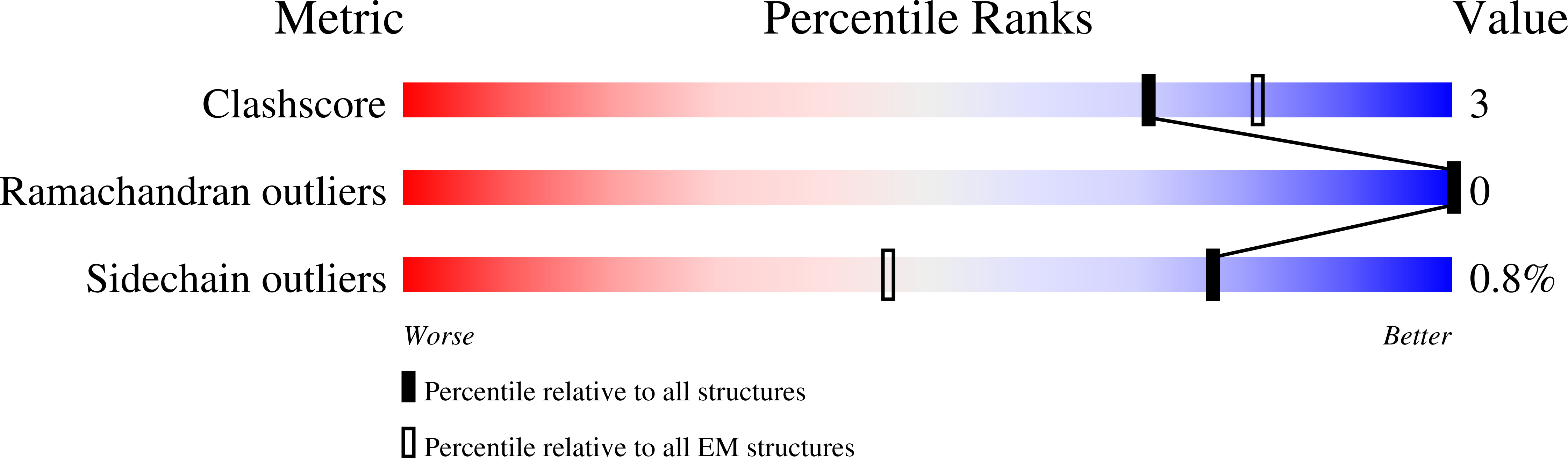
Deposition Date
2024-03-27
Release Date
2025-05-07
Last Version Date
2025-10-08
Entry Detail
PDB ID:
9B7G
Keywords:
Title:
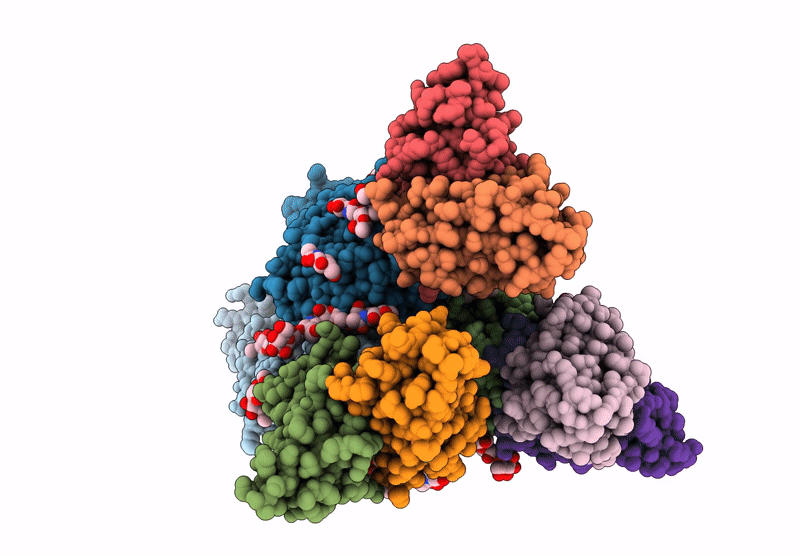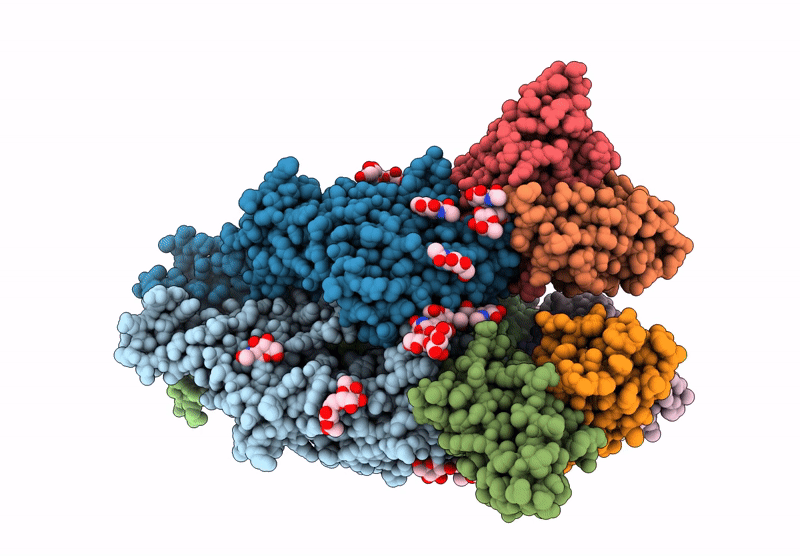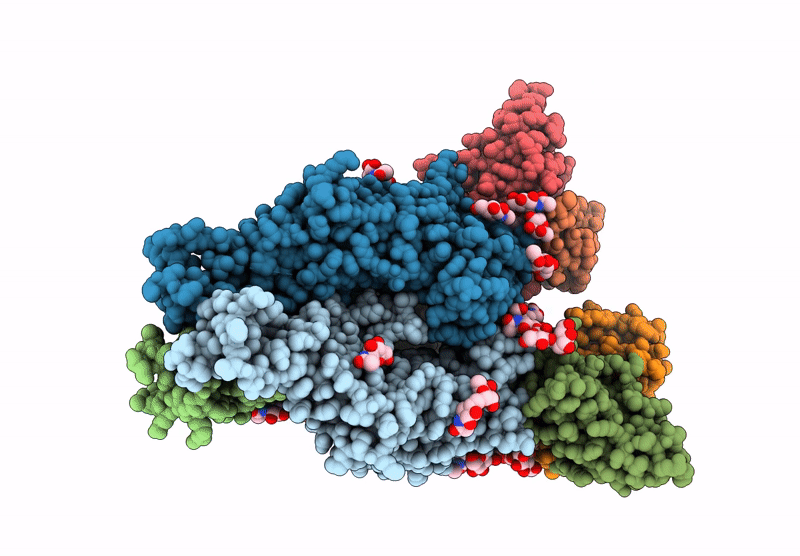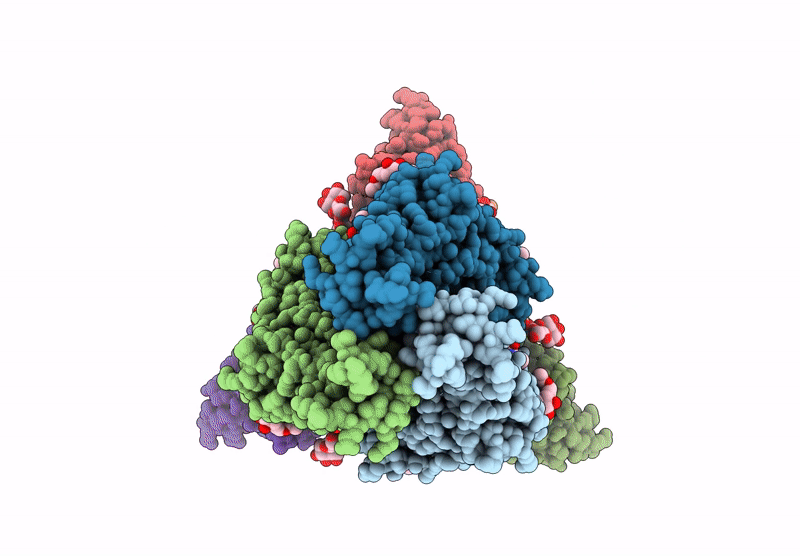
Cryo-EM structure of antibody TJ5-13 bound to H3 COBRA NG2 hemagglutinin
Biological Source:
Source Organism(s):
Influenza A virus (Taxon ID: 11320)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.61 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE