Deposition Date
2024-03-20
Release Date
2025-03-12
Last Version Date
2025-07-23
Entry Detail
PDB ID:
9B4I
Keywords:
Title:
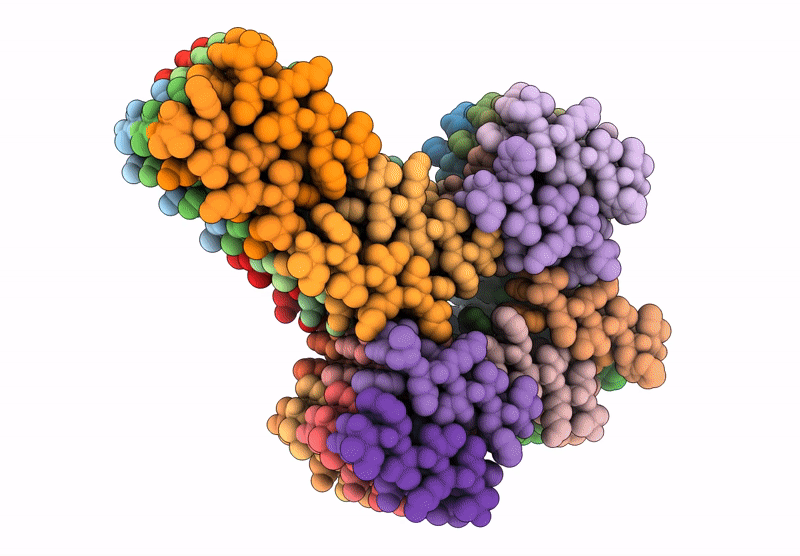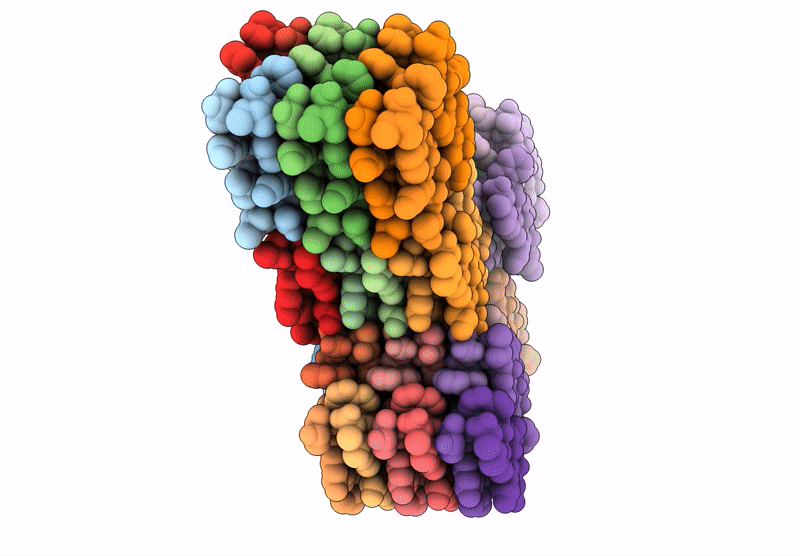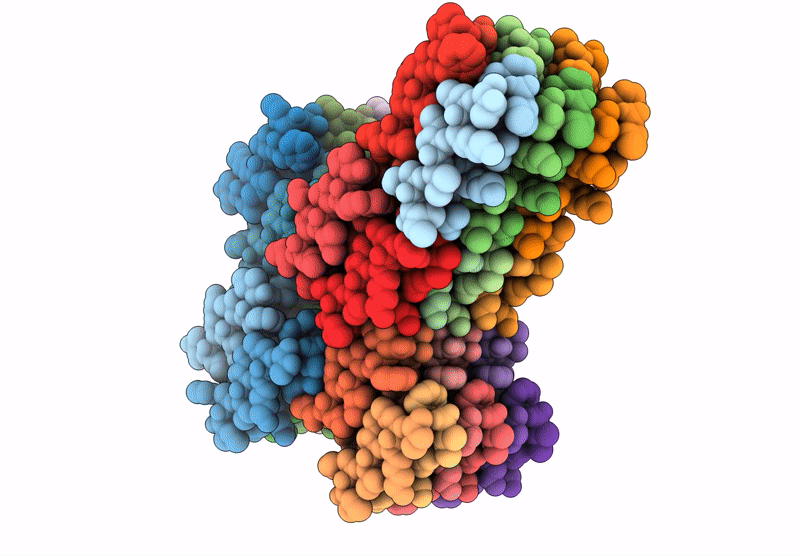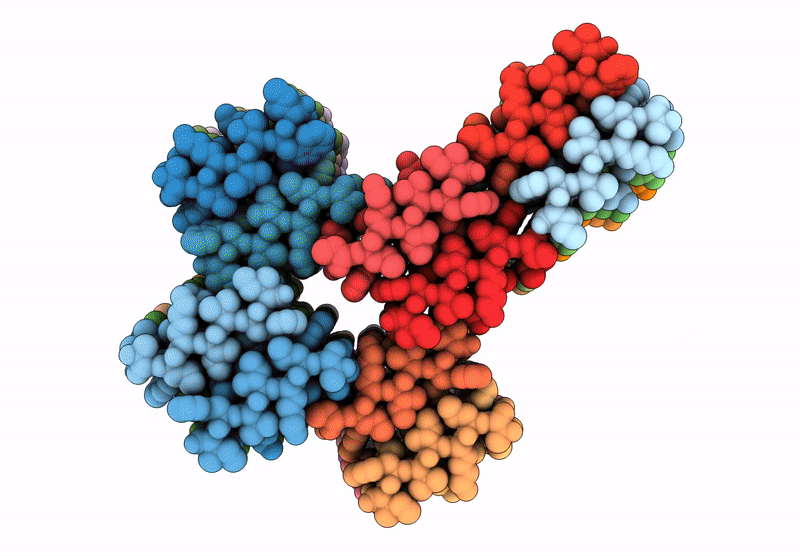
Filament of D-TLKIVWI, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Resolution:
3.60 Å
Aggregation State:
FILAMENT
Reconstruction Method:
HELICAL


