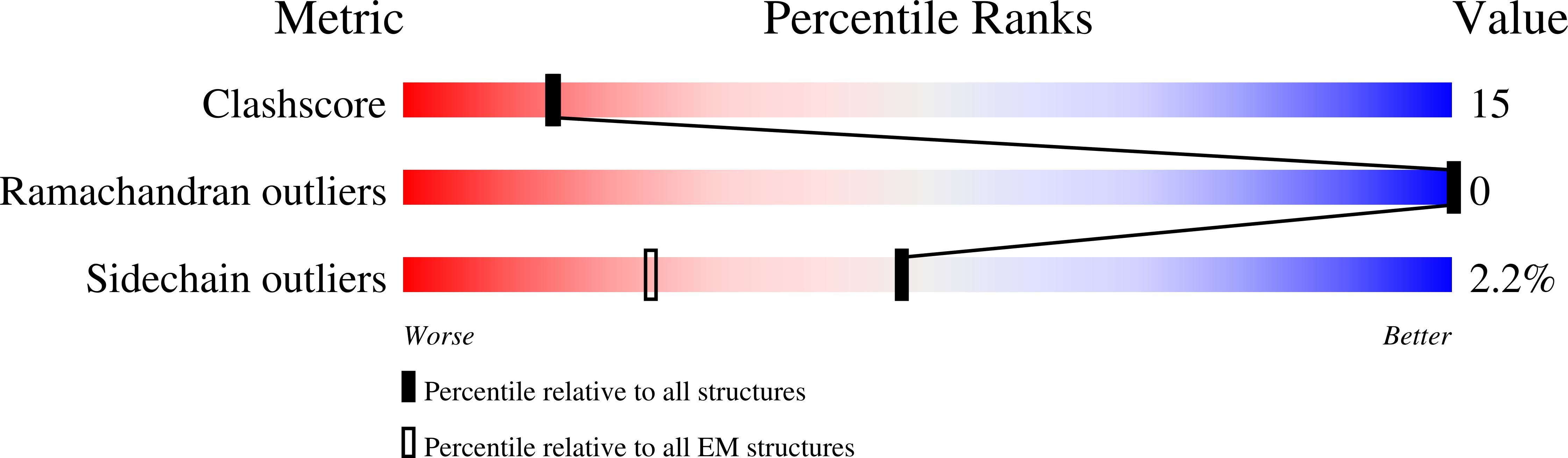
Deposition Date
2024-03-19
Release Date
2025-04-23
Last Version Date
2025-06-04
Entry Detail
PDB ID:
9B3H
Keywords:
Title:
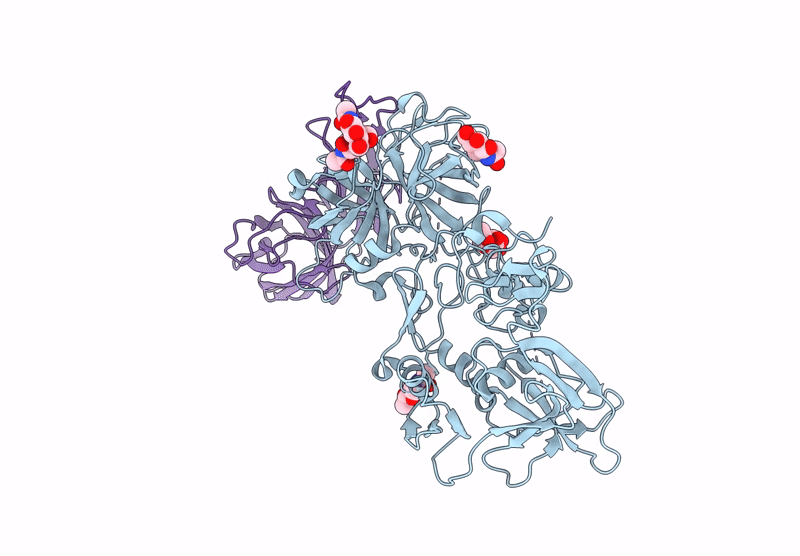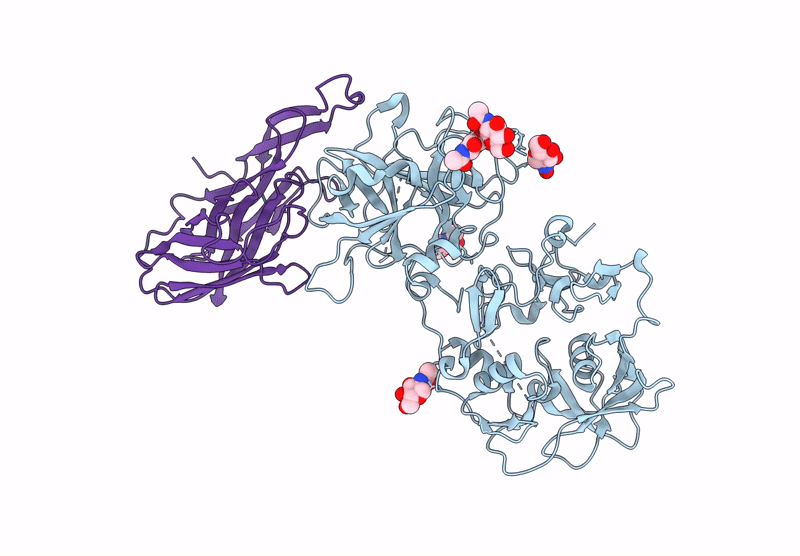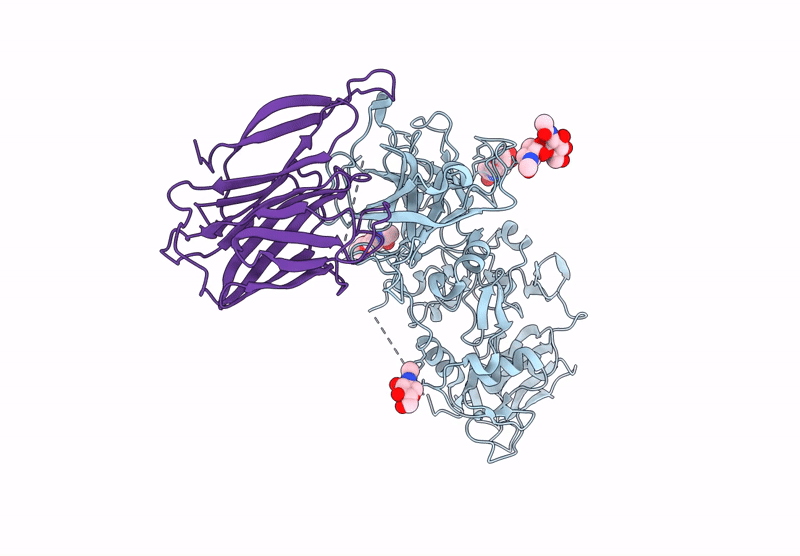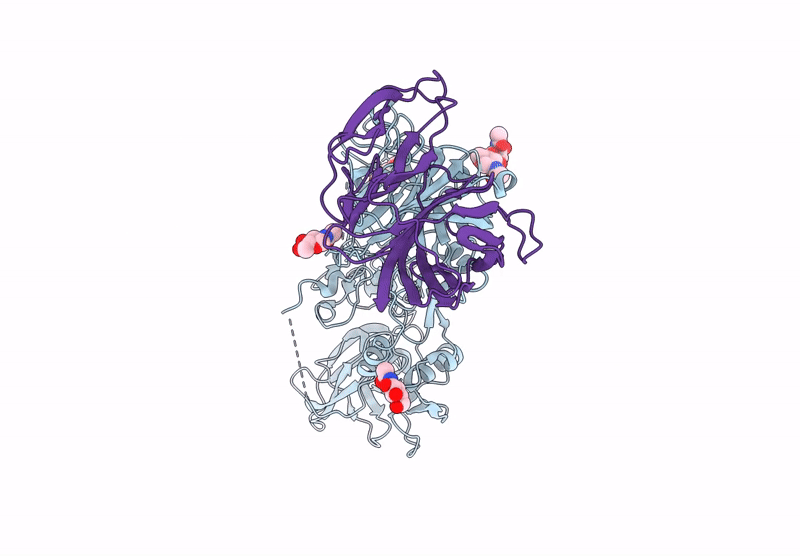
Structure of a complex between Pasteurella multocida surface lipoprotein, PmSLP-1, and bovine complement factor I
Biological Source:
Source Organism(s):
Pasteurella multocida 36950 (Taxon ID: 1075089)
Bos taurus (Taxon ID: 9913)
Bos taurus (Taxon ID: 9913)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE