Deposition Date
2024-06-16
Release Date
2025-02-05
Last Version Date
2025-07-16
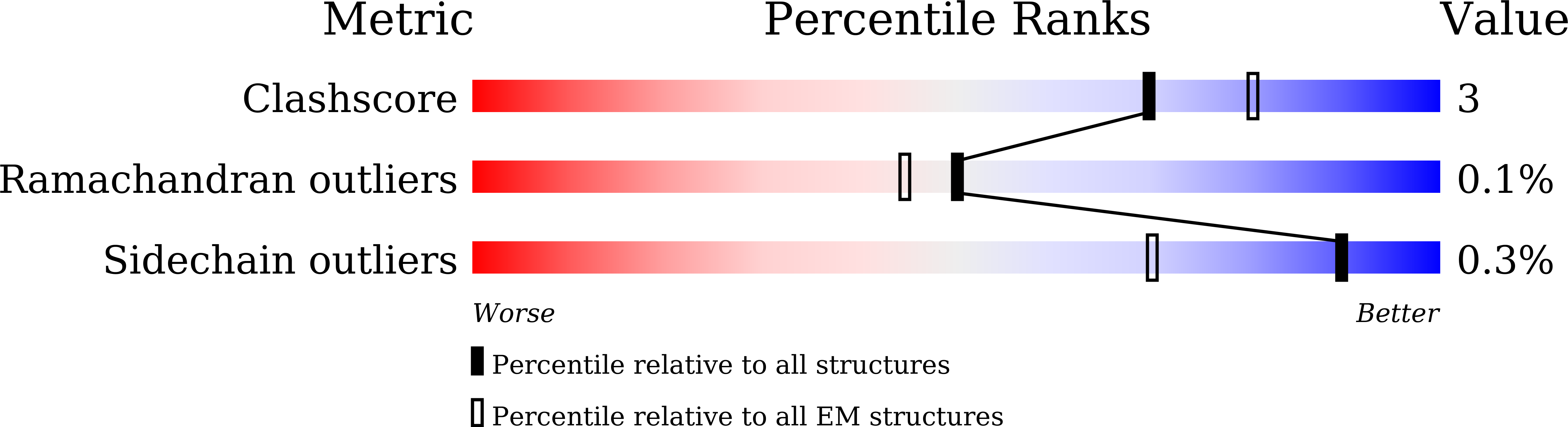
Entry Detail
PDB ID:
8ZY9
Keywords:
Title:
Ra9479 Bat ACE2 Dimer in Complex with Two BtKY72 Sarbecovirus Spike RBDs.
Biological Source:
Source Organism:
Kenya bat coronavirus BtKY72 (Taxon ID: 903610)
Rhinolophus affinis (Taxon ID: 59477)
Rhinolophus affinis (Taxon ID: 59477)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE