Deposition Date
2024-04-23
Release Date
2025-04-30
Last Version Date
2025-05-14
Entry Detail
PDB ID:
8Z9I
Keywords:
Title:
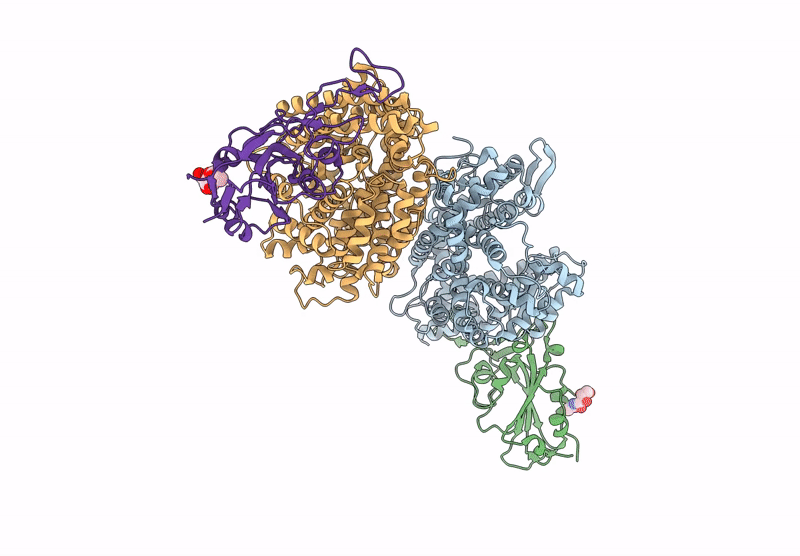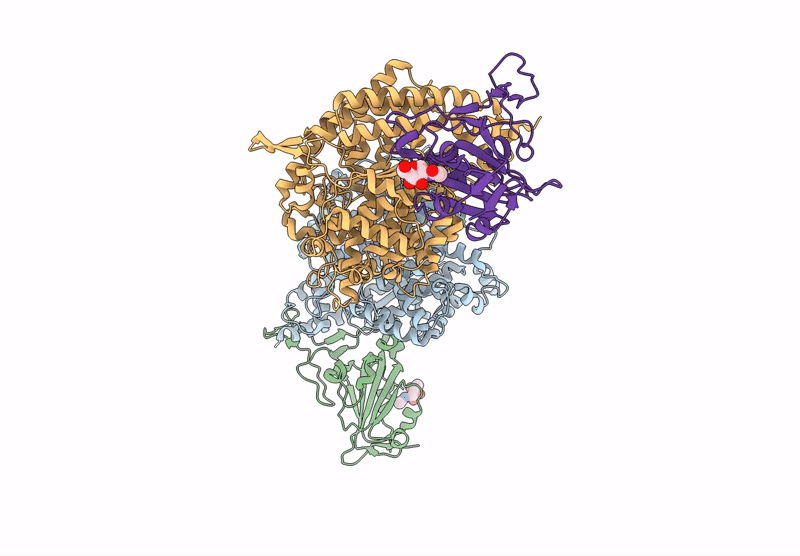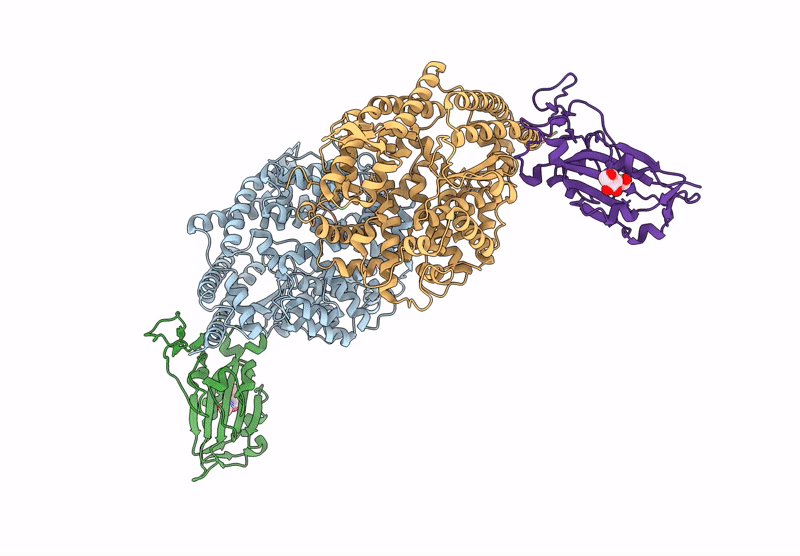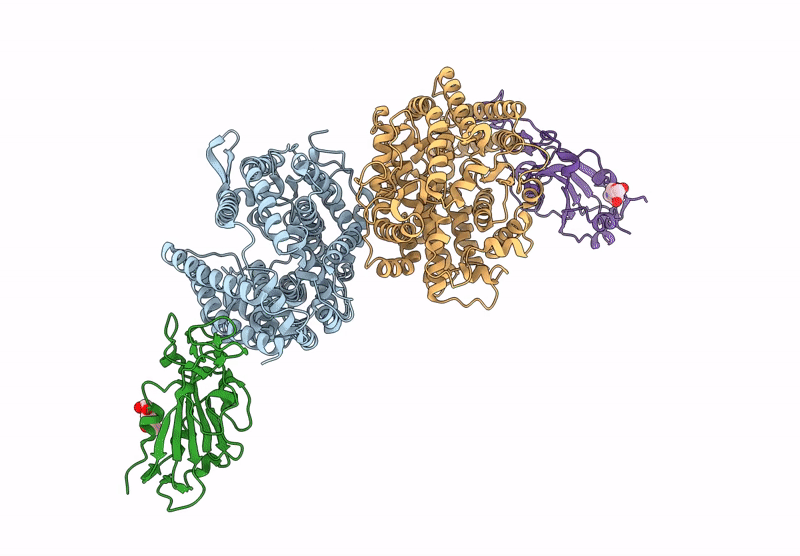
Crystal structure of RaTG13 RBD bound to Rhinolophus affinis ACE2
Biological Source:
Source Organism:
Bat coronavirus RaTG13 (Taxon ID: 2709072)
Rhinolophus affinis (Taxon ID: 59477)
Rhinolophus affinis (Taxon ID: 59477)
Host Organism:
Method Details:
Experimental Method:
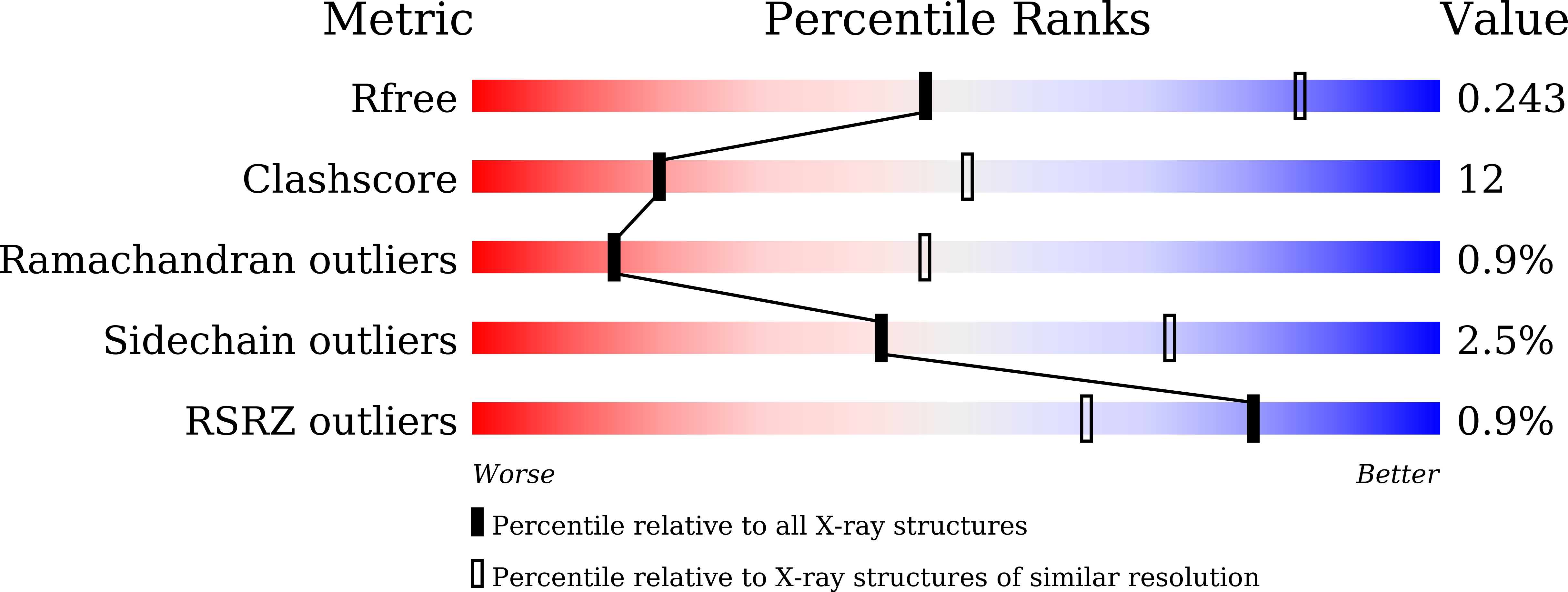
Resolution:
3.01 Å
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
C 2 2 21