Deposition Date
2024-04-15
Release Date
2024-12-11
Last Version Date
2025-07-16
Entry Detail
PDB ID:
8Z3K
Keywords:
Title:
The structure of type III CRISPR-associated deaminase in complex 2cA6-2ATP
Biological Source:
Source Organism(s):
Limisphaera ngatamarikiensis (Taxon ID: 1324935)
Expression System(s):
Method Details:
Experimental Method:
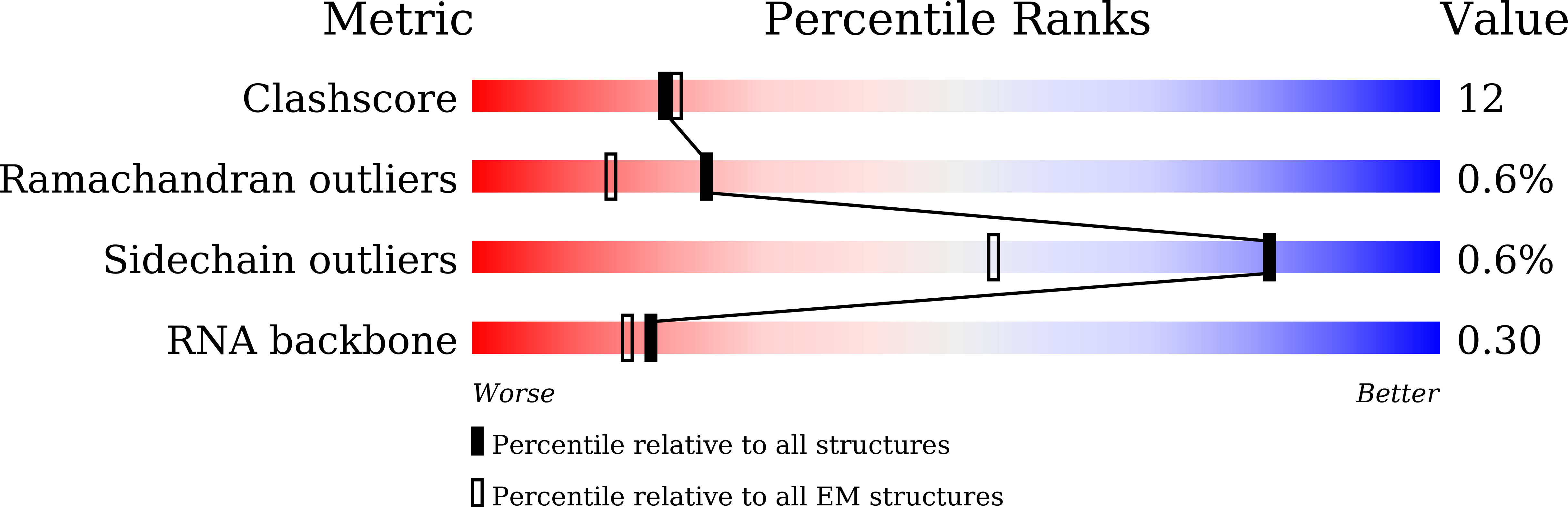
Resolution:
3.19 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE