Deposition Date
2024-02-27
Release Date
2025-03-05
Last Version Date
2025-09-17
Entry Detail
PDB ID:
8YH1
Keywords:
Title:
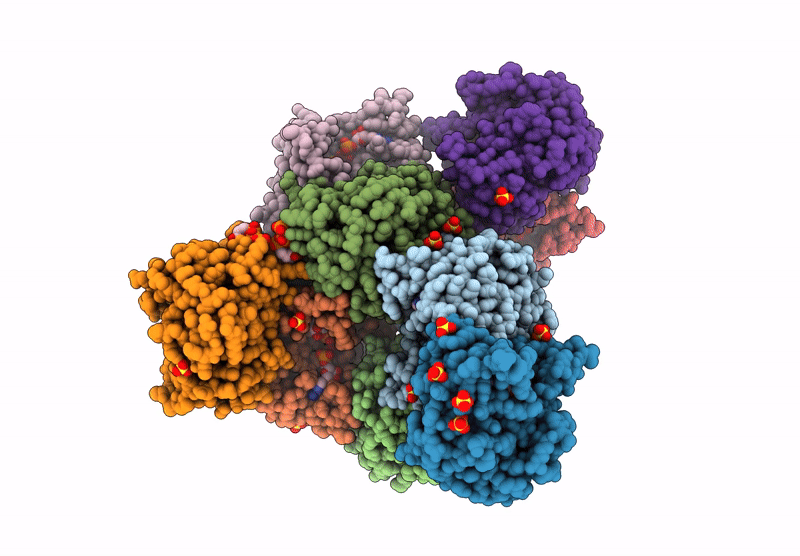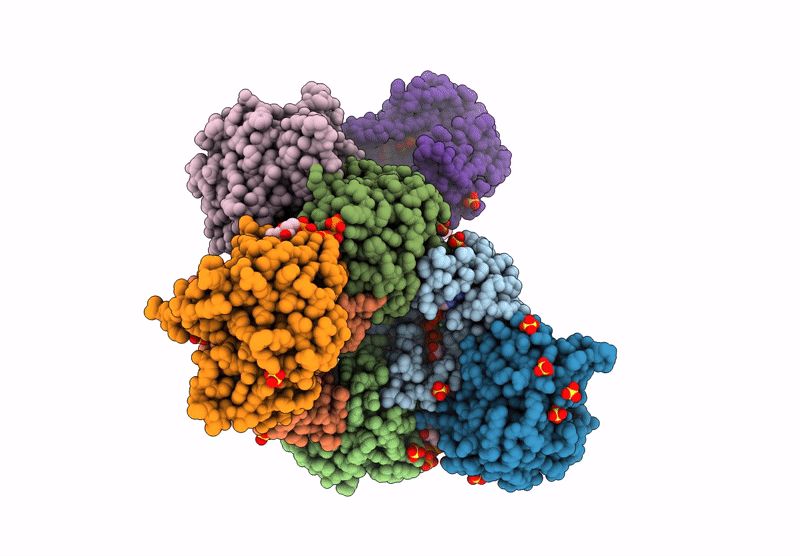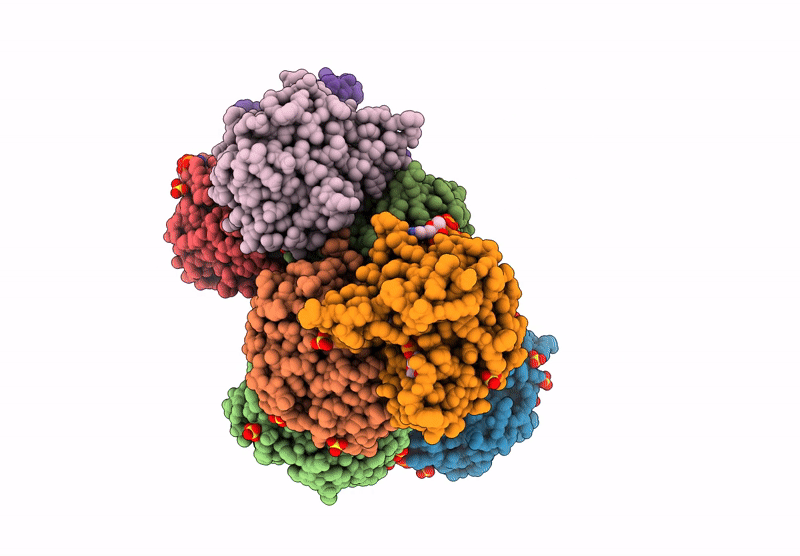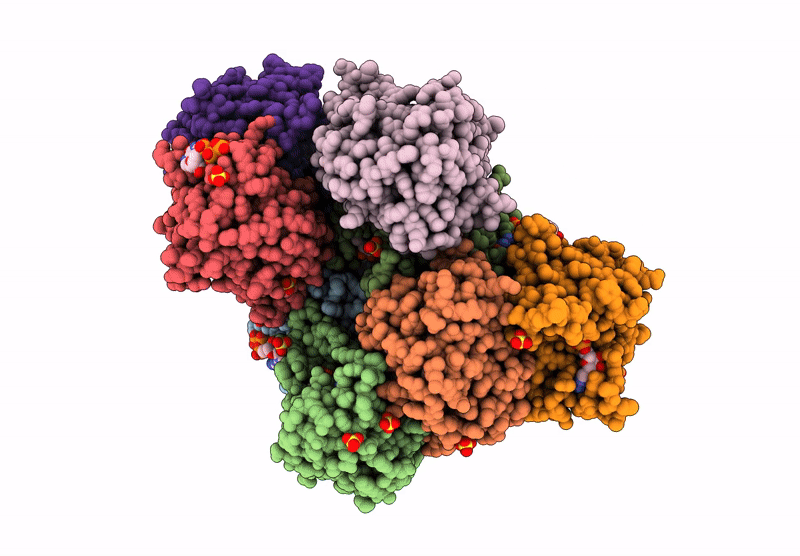
Crystal structure of Thermus thermophilus UMP kinase complexed with a phosphoryl group acceptor and donor.
Biological Source:
Source Organism:
Thermus thermophilus HB8 (Taxon ID: 300852)
Host Organism:
Method Details:
Experimental Method:
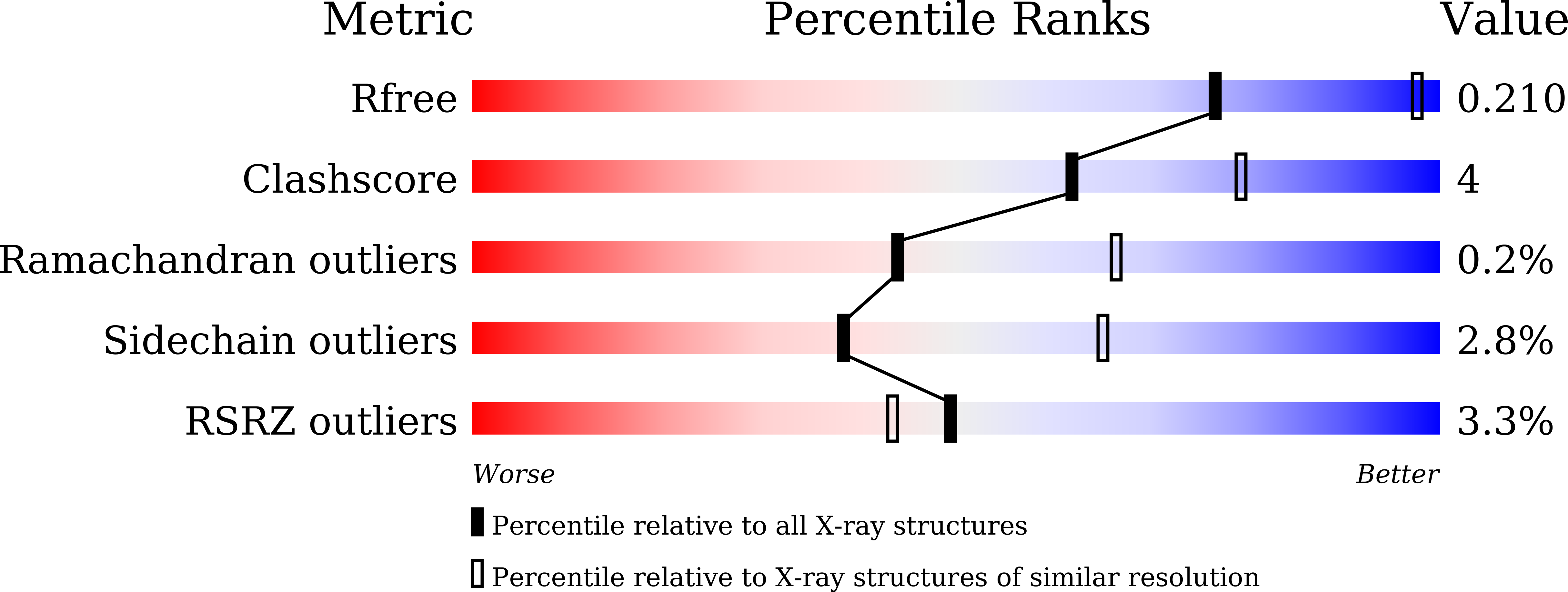
Resolution:
2.60 Å
R-Value Free:
0.20
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
I 2 2 2