Deposition Date
2023-12-08
Release Date
2024-12-04
Last Version Date
2024-12-11
Entry Detail
PDB ID:
8XC7
Keywords:
Title:
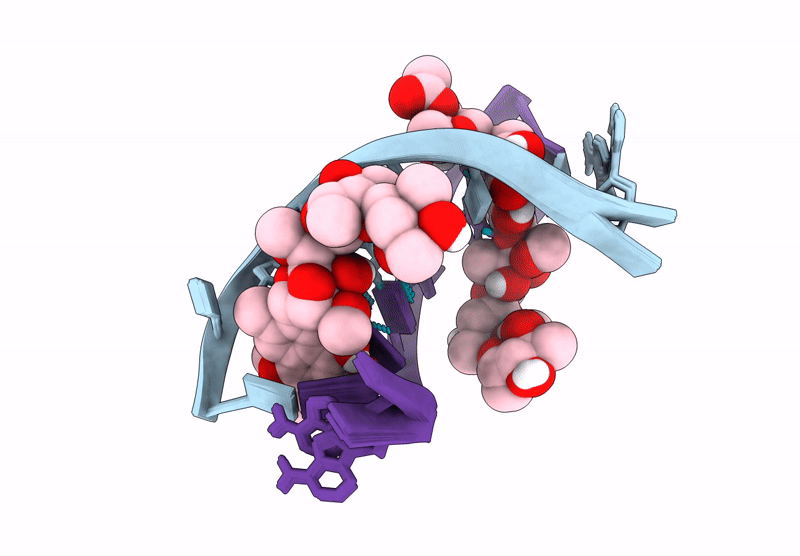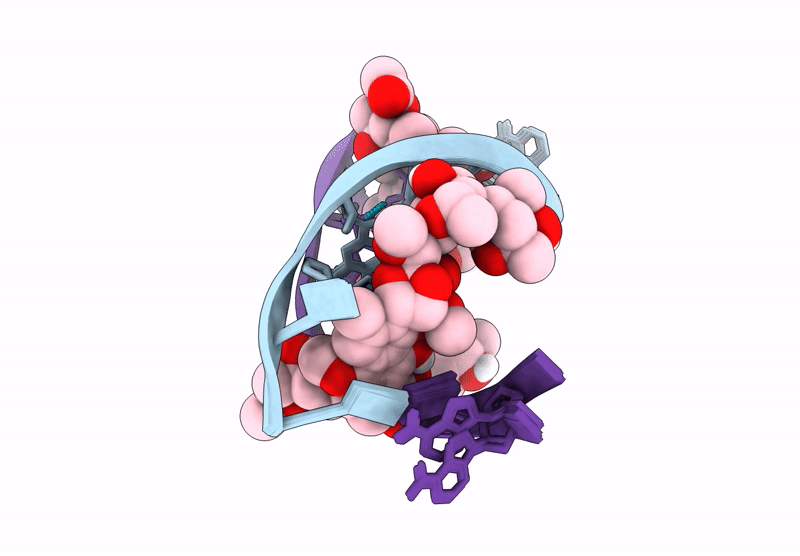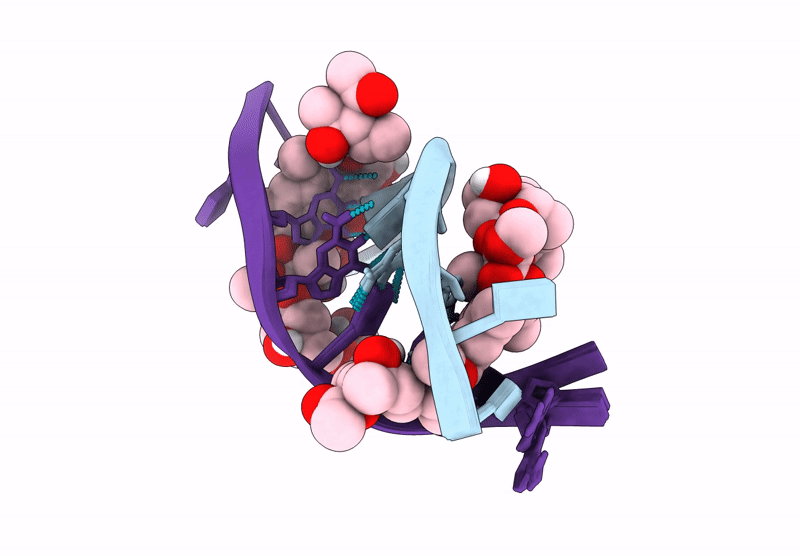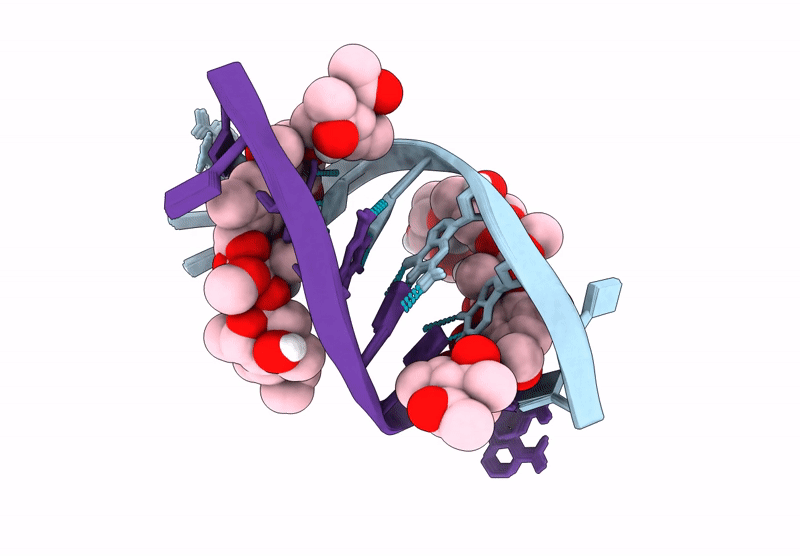
Structure of LL-D49194-alpha-1 covalently bound to guanosine-2'-fluorinated d(AACCGGTT)2
Biological Source:
Source Organism(s):
DNA molecule (Taxon ID: 2853804)
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy


