Deposition Date
2023-10-18
Release Date
2024-06-26
Last Version Date
2024-11-06
Entry Detail
PDB ID:
8WT6
Keywords:
Title:
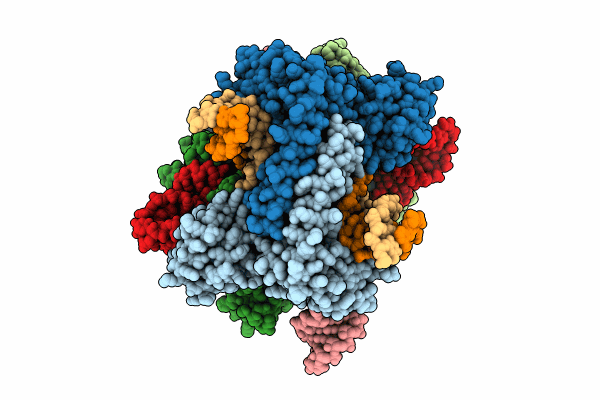
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
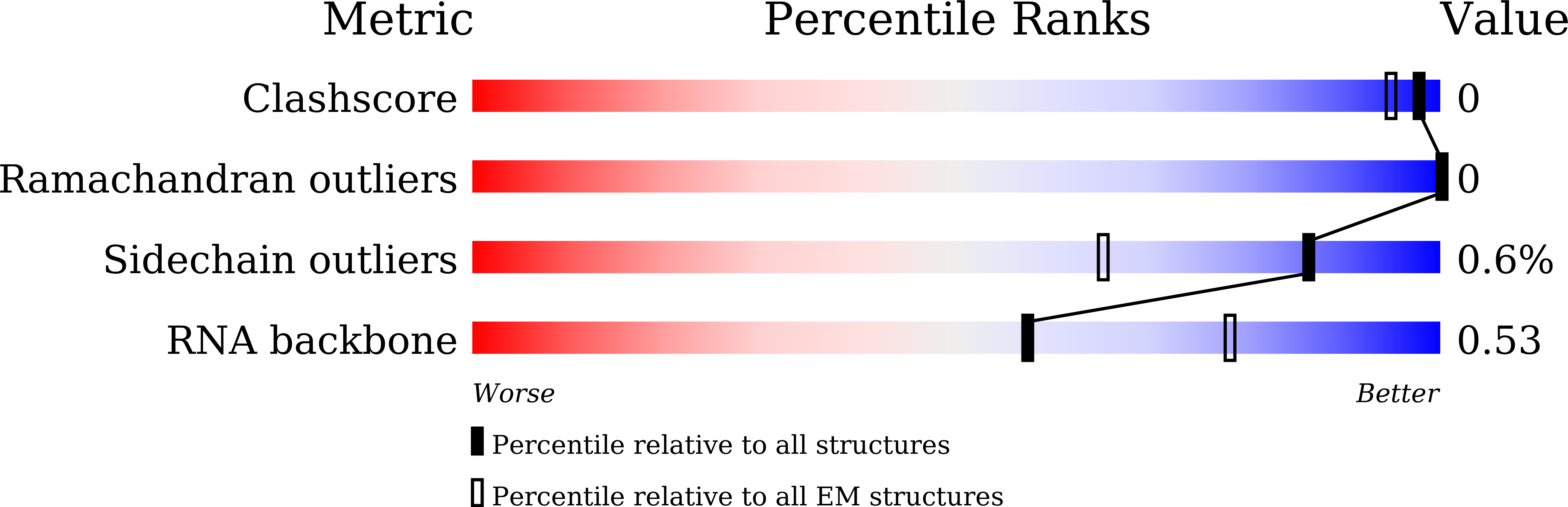
Resolution:
2.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE