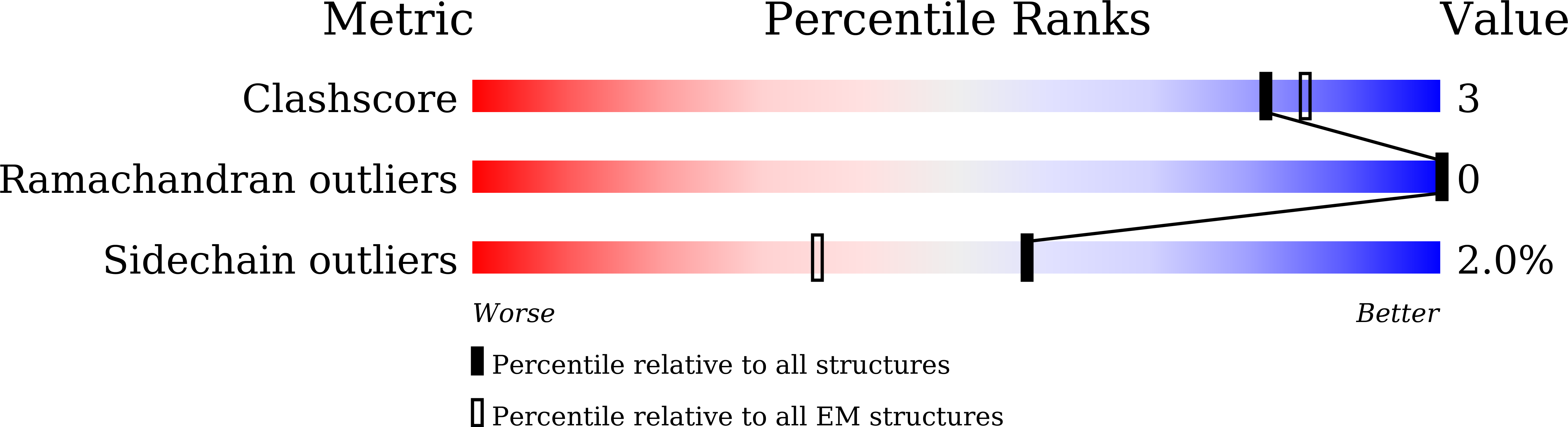
Deposition Date
2023-10-09
Release Date
2023-12-27
Last Version Date
2023-12-27
Entry Detail
PDB ID:
8WP9
Keywords:
Title:
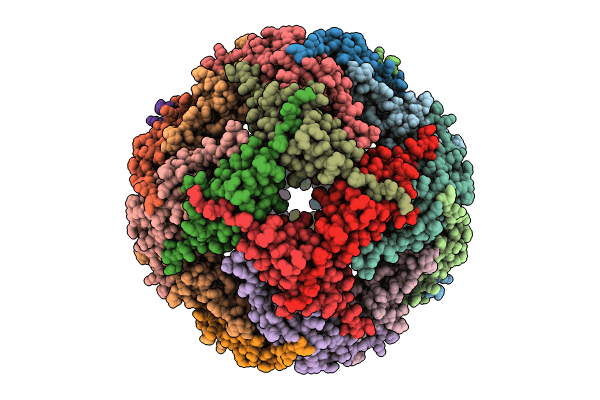
Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5
Biological Source:
Source Organism(s):
Methanocaldococcus jannaschii DSM 2661 (Taxon ID: 243232)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.49 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE