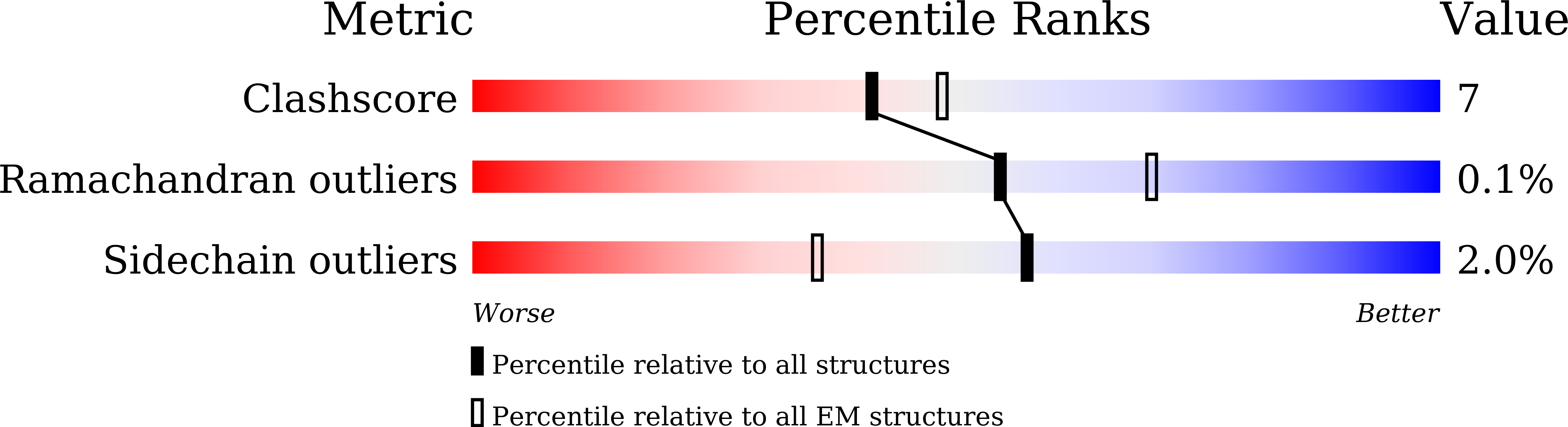
Deposition Date
2023-10-03
Release Date
2024-05-29
Last Version Date
2024-11-13
Entry Detail
PDB ID:
8WM6
Keywords:
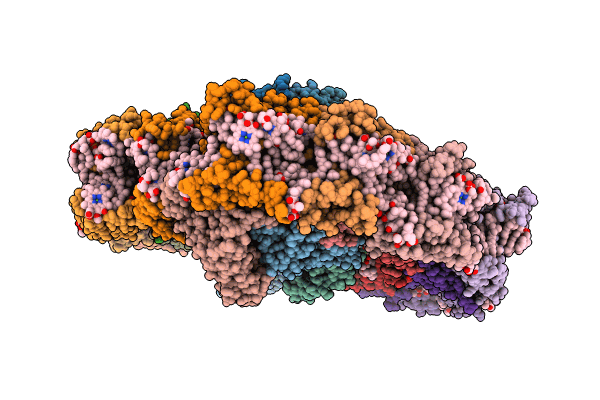
Title:
The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution
Biological Source:
Source Organism(s):
Rhodomonas salina (Taxon ID: 52970)
Method Details:
Experimental Method:
Resolution:
2.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE