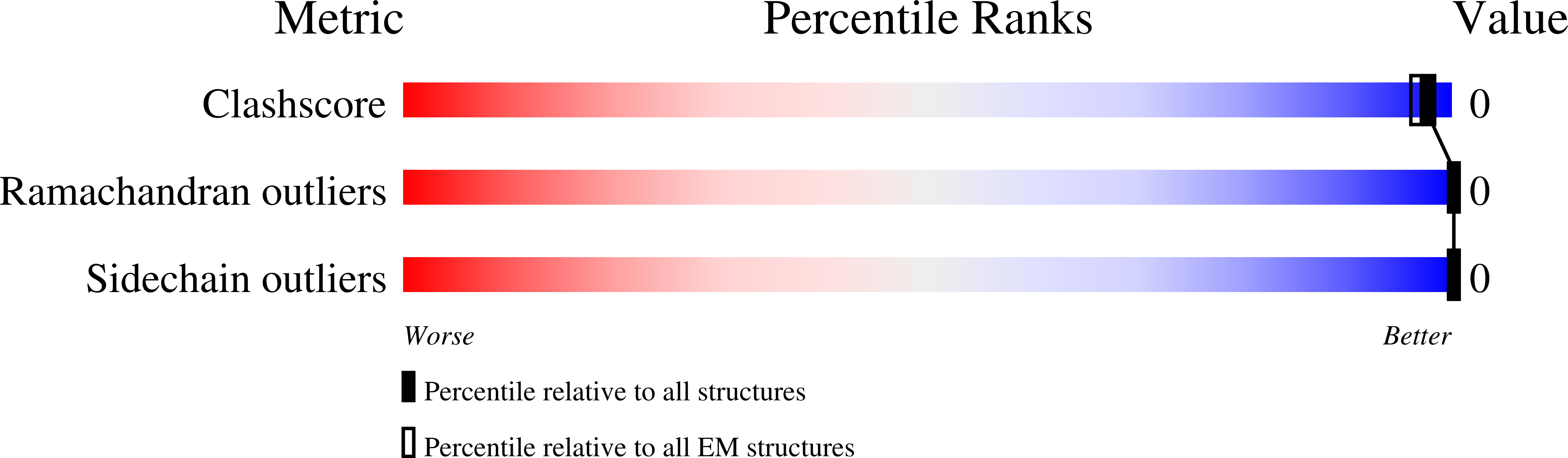
Deposition Date
2024-01-16
Release Date
2025-03-19
Last Version Date
2025-06-04
Entry Detail
PDB ID:
8VPF
Keywords:
Title:
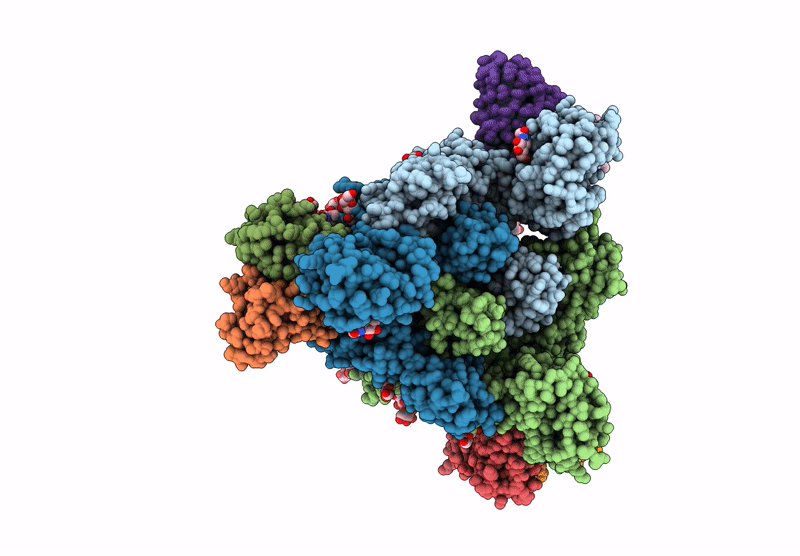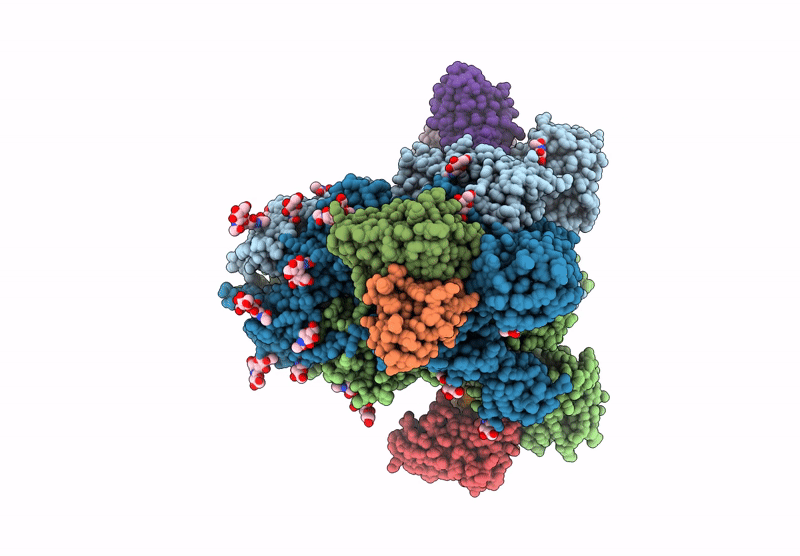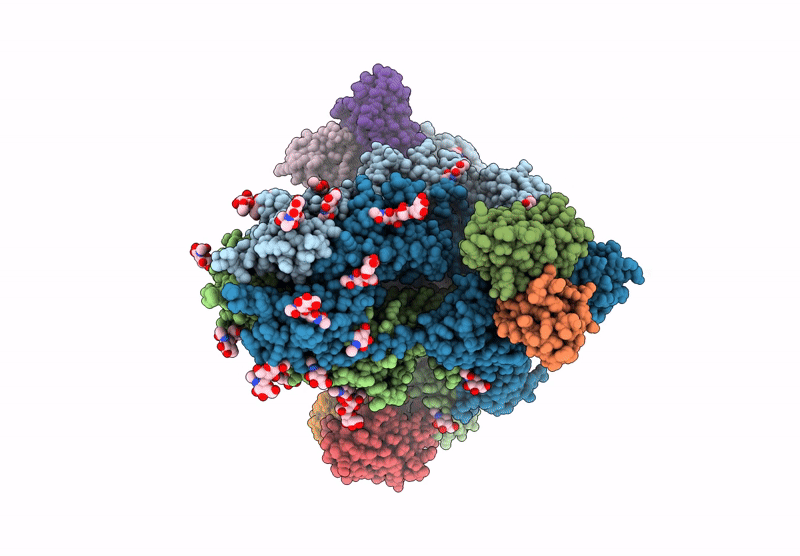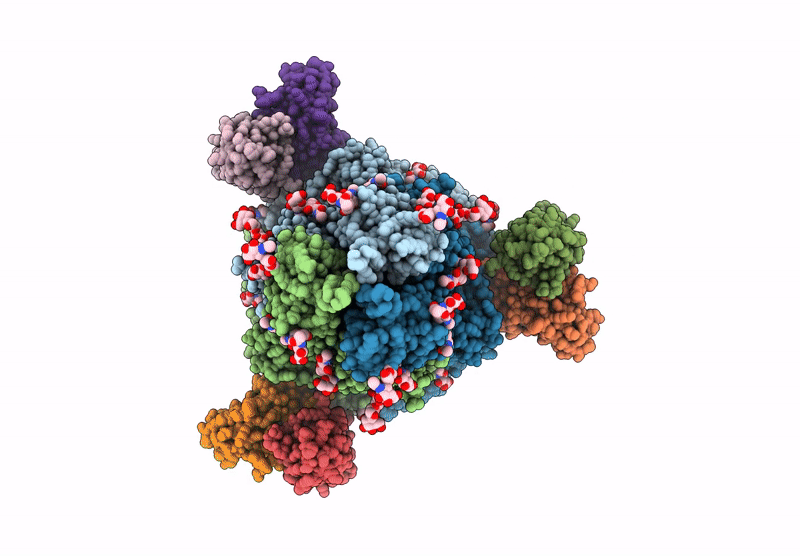
Structure of SARS-CoV spike in complex with CoV1-65 Fab (NTD-bound)
Biological Source:
Source Organism(s):
Severe acute respiratory syndrome coronavirus (Taxon ID: 2901879)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE