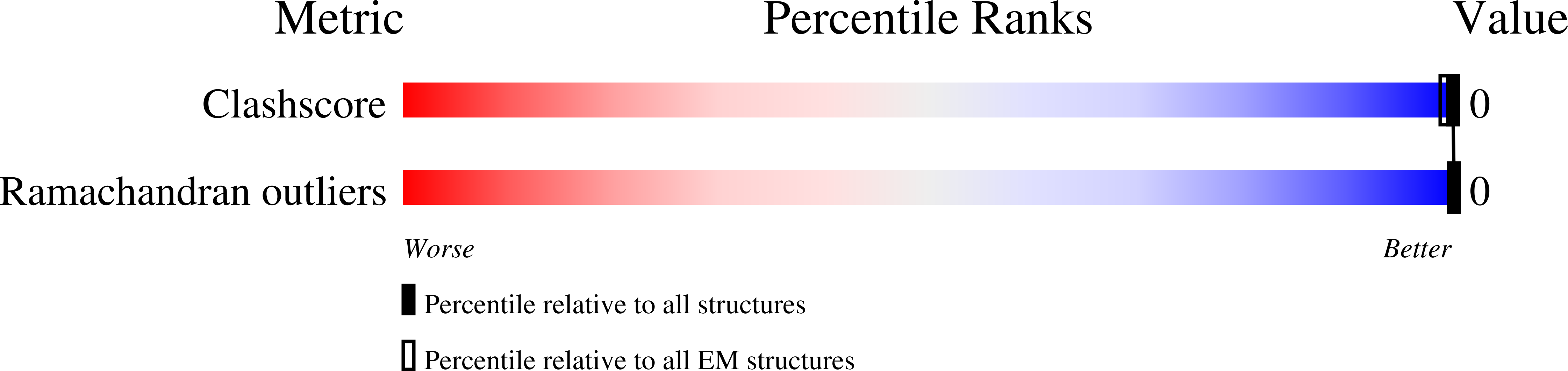
Deposition Date
2023-10-20
Release Date
2024-08-14
Last Version Date
2024-09-11
Entry Detail
PDB ID:
8UP1
Keywords:
Title:
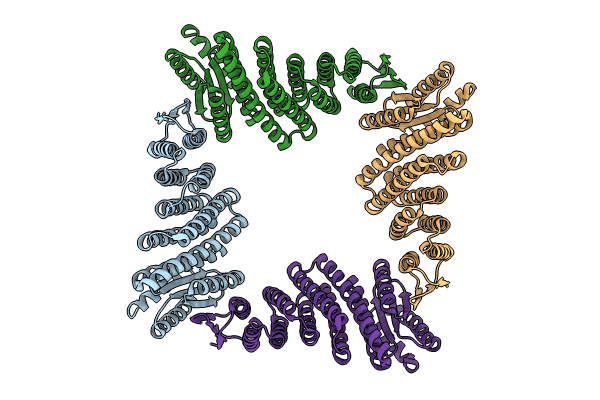
CryoEM Structure of Allosterically Switchable De Novo Protein sr322, In Closed State without Effector Peptide
Biological Source:
Source Organism(s):
unidentified (Taxon ID: 32644)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.55 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE