Deposition Date
2023-10-11
Release Date
2024-03-06
Last Version Date
2024-06-19
Entry Detail
PDB ID:
8UJA
Keywords:
Title:
T33-fn10 - Designed Tetrahedral Protein Cage Using Fragment-based Hydrogen Bond Networks
Biological Source:
Source Organism(s):
Sulfurisphaera tokodaii str. 7 (Taxon ID: 273063)
Novosphingobium aromaticivorans DSM 12444 (Taxon ID: 279238)
Novosphingobium aromaticivorans DSM 12444 (Taxon ID: 279238)
Expression System(s):
Method Details:
Experimental Method:
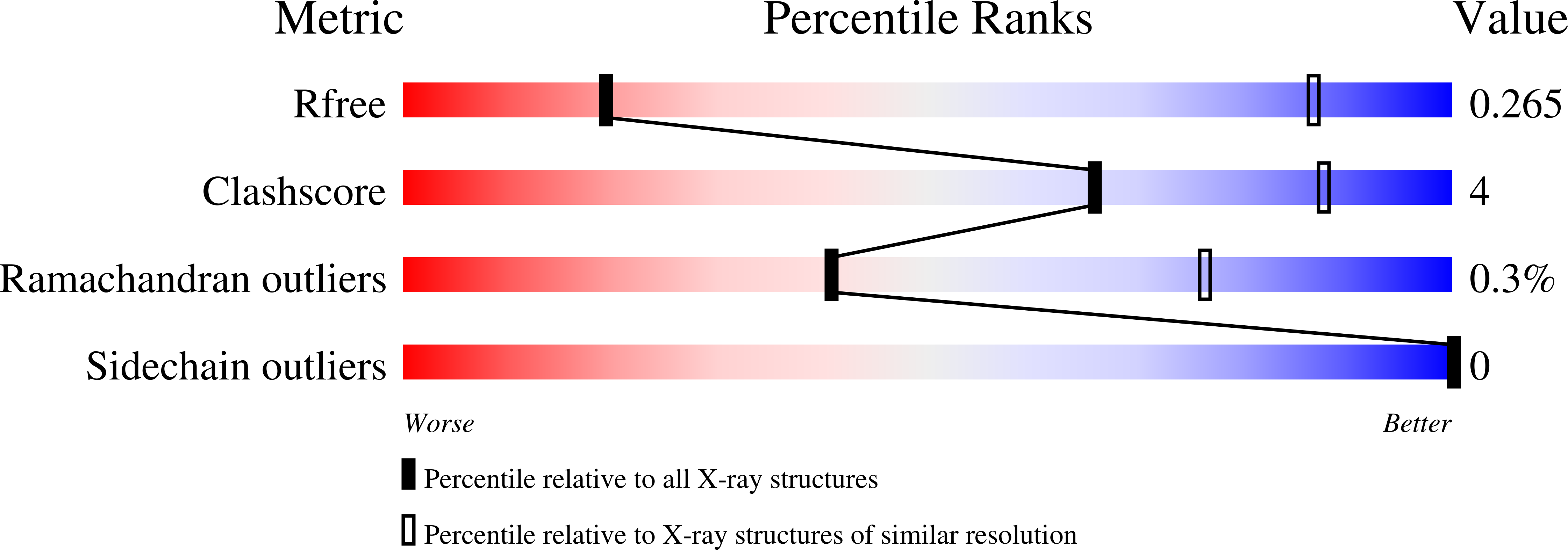
Resolution:
6.00 Å
R-Value Free:
0.24
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 3