Deposition Date
2023-08-18
Release Date
2024-03-27
Last Version Date
2024-04-10
Entry Detail
PDB ID:
8TW1
Keywords:
Title:
Crystal structure of Lys2972, a phage endolysin targeting Streptococcus thermophilus
Biological Source:
Source Organism:
Streptococcus phage 2972 (Taxon ID: 2908019)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.27 Å
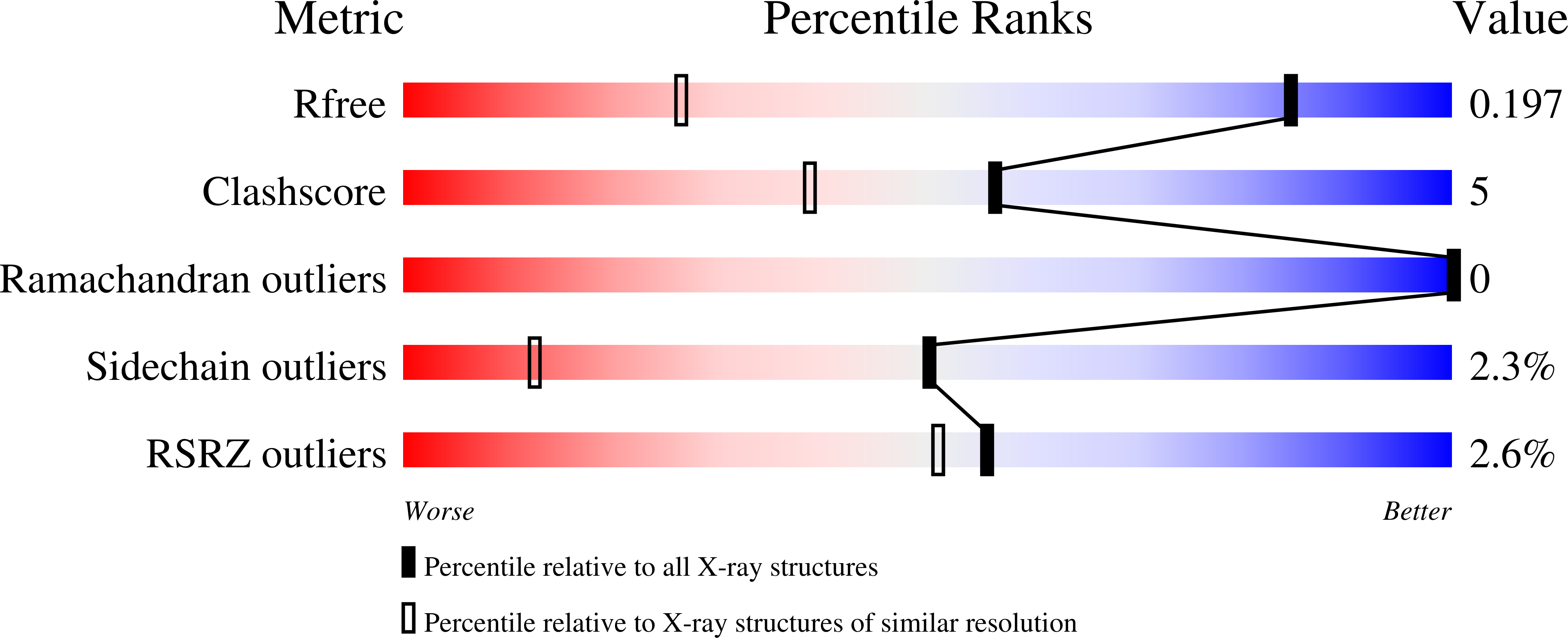
R-Value Free:
0.19
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
C 1 2 1