Deposition Date
2023-08-04
Release Date
2024-06-19
Last Version Date
2024-10-23
Entry Detail
PDB ID:
8TOW
Keywords:
Title:
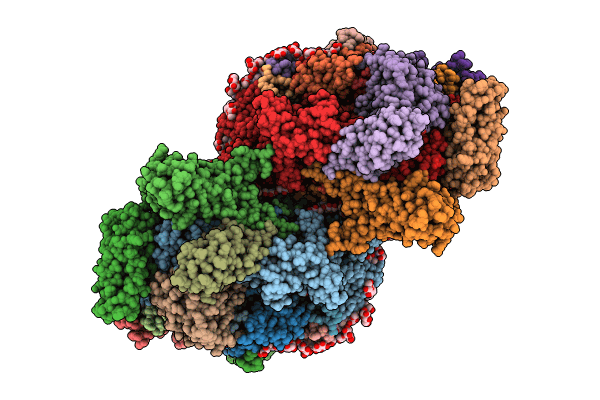
Structure of a mutated photosystem II complex reveals perturbation of the oxygen-evolving complex
Biological Source:
Source Organism(s):
Synechocystis sp. PCC 6803 (Taxon ID: 1148)
Method Details:
Experimental Method:
Resolution:
2.14 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE