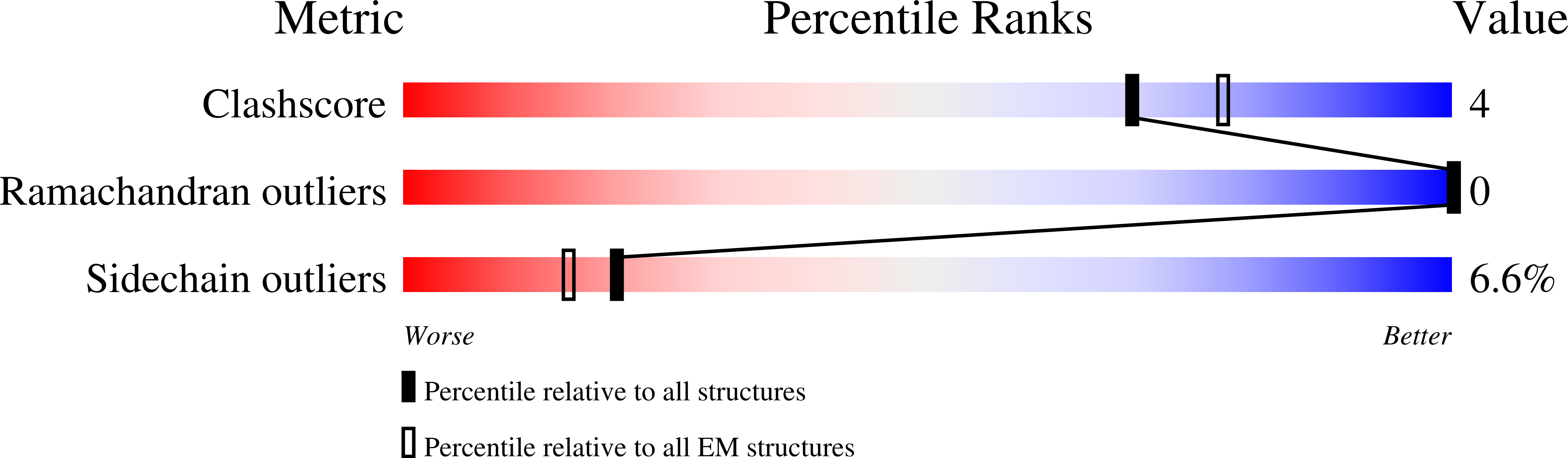
Deposition Date
2023-07-25
Release Date
2023-09-13
Last Version Date
2023-09-20
Entry Detail
PDB ID:
8TK7
Keywords:
Title:
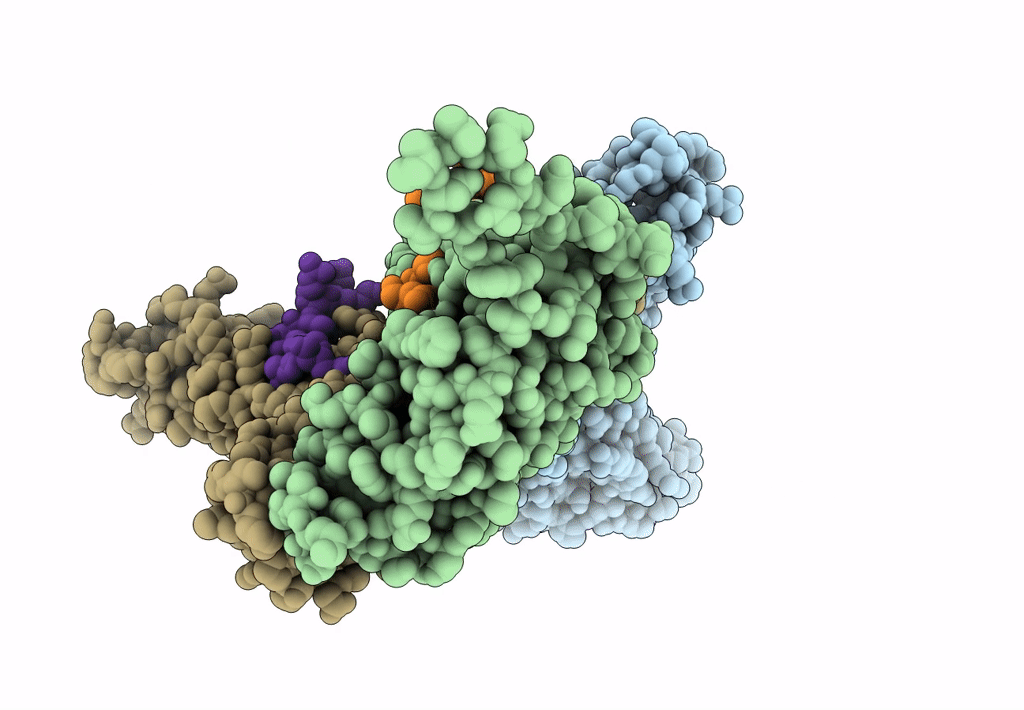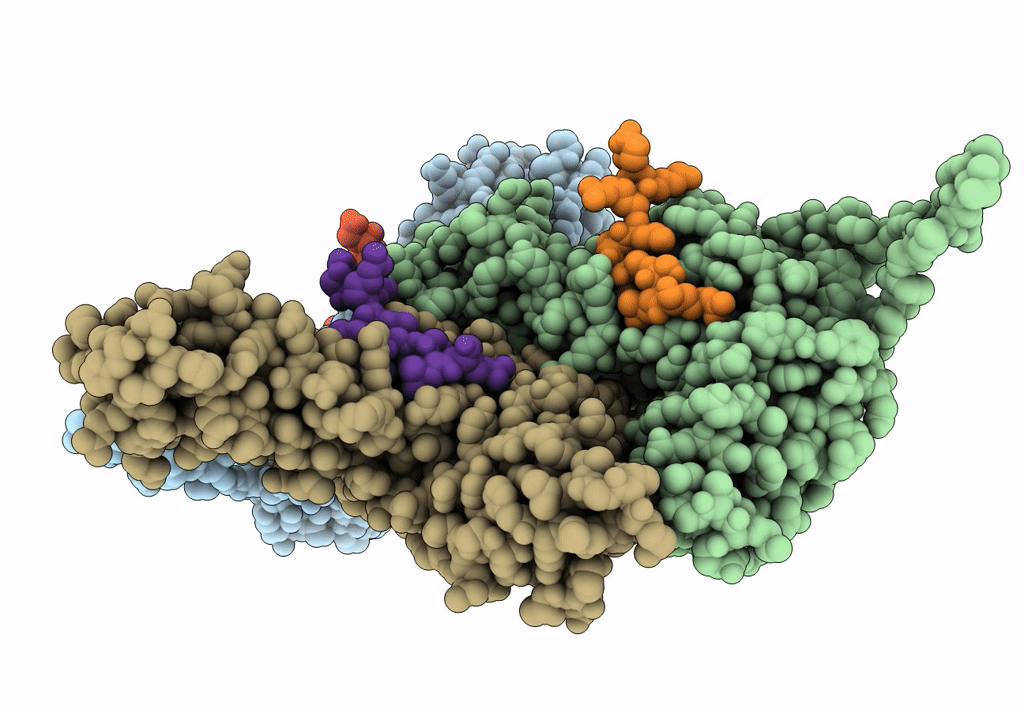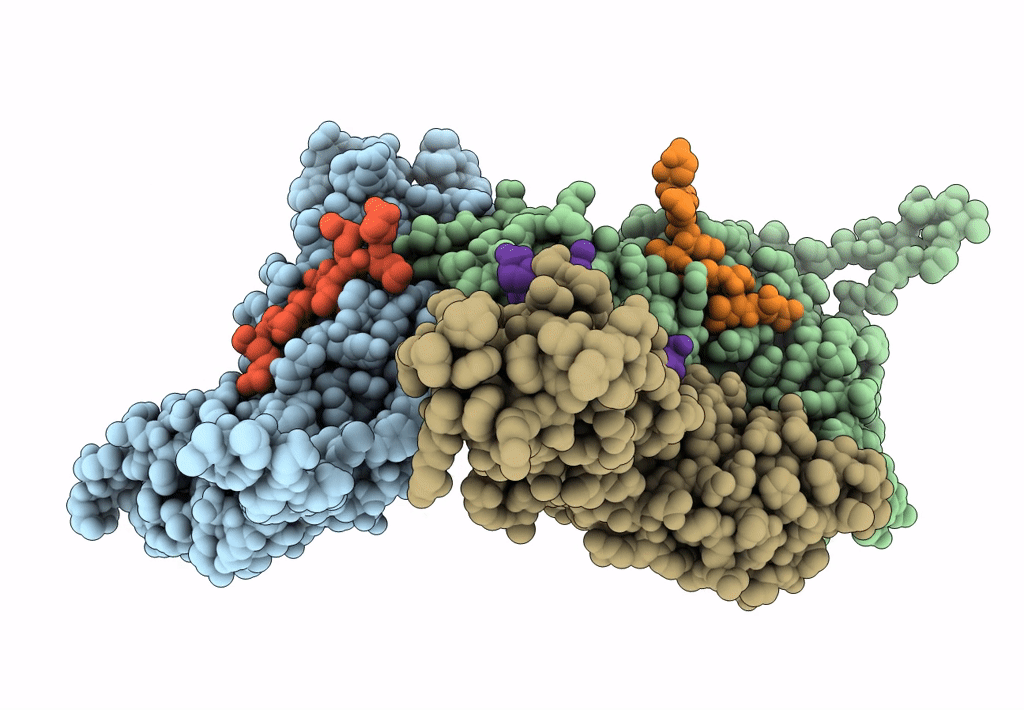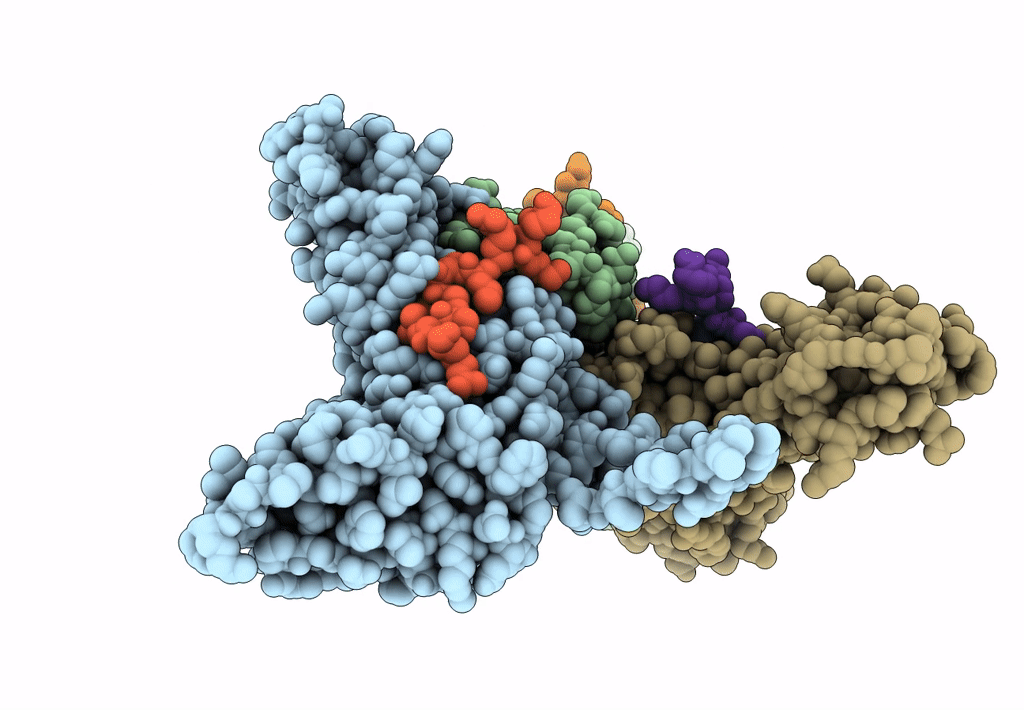
Myxococcus xanthus EncA protein shell with compartmentalized SNAP-tag cargo protein
Biological Source:
Source Organism(s):
Myxococcus xanthus DK 1622 (Taxon ID: 246197)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.53 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE