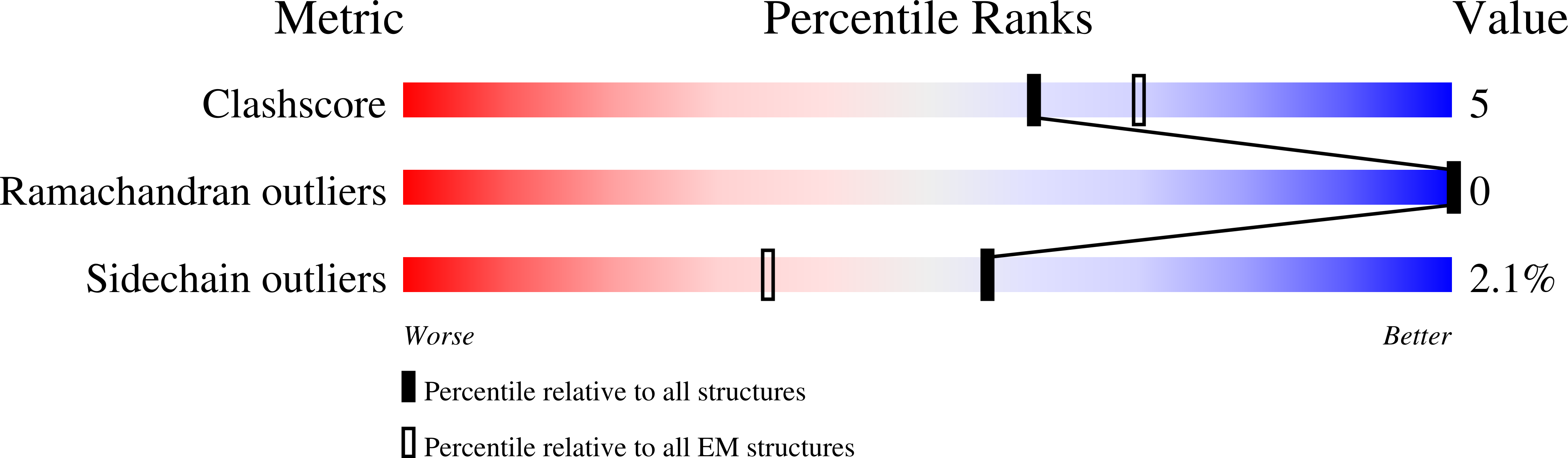
Deposition Date
2023-07-16
Release Date
2023-11-22
Last Version Date
2024-02-28
Entry Detail
PDB ID:
8THJ
Keywords:
Title:
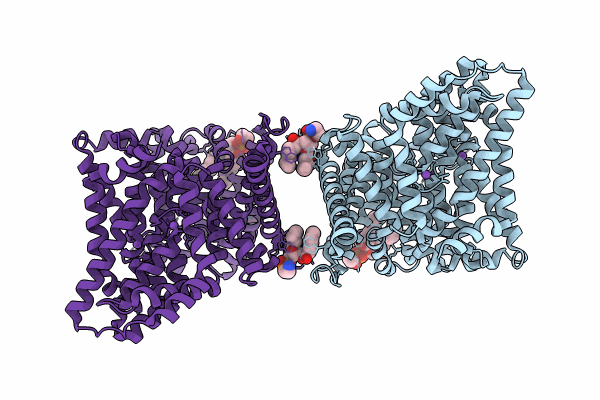
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer)
Biological Source:
Source Organism(s):
Haemophilus influenzae Rd KW20 (Taxon ID: 71421)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.99 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE