Deposition Date
2023-06-28
Release Date
2023-10-25
Last Version Date
2025-05-14
Entry Detail
PDB ID:
8TAZ
Keywords:
Title:
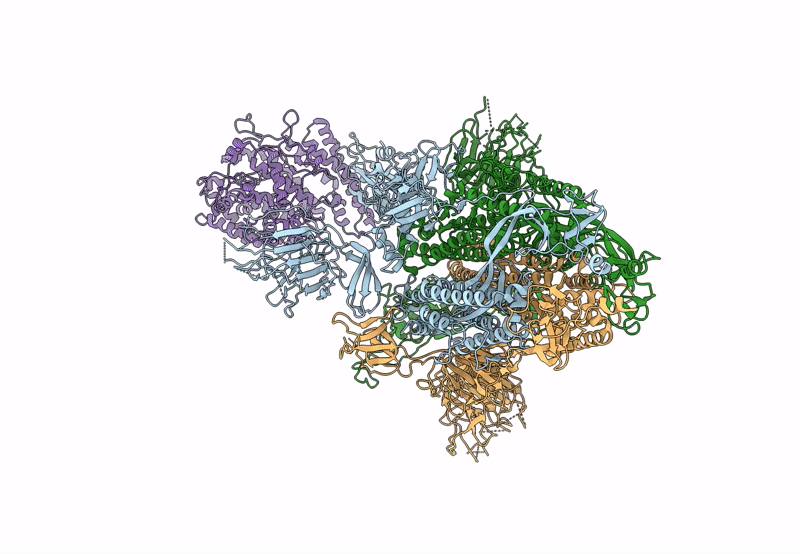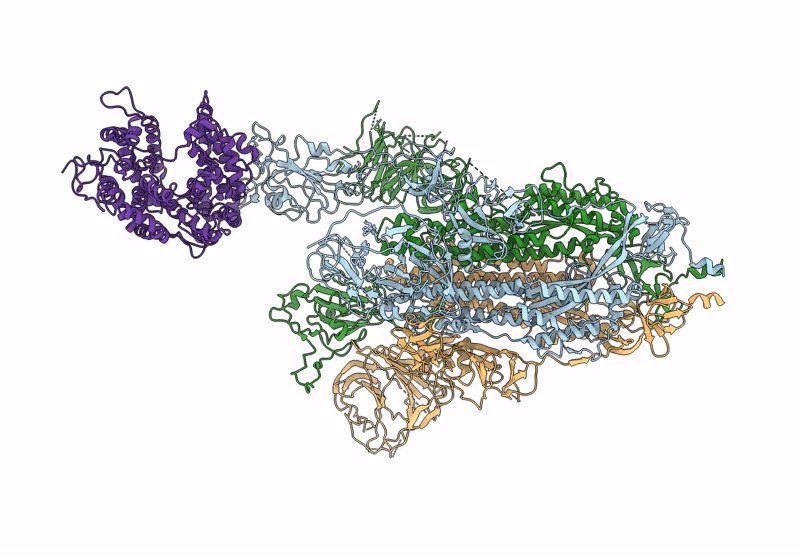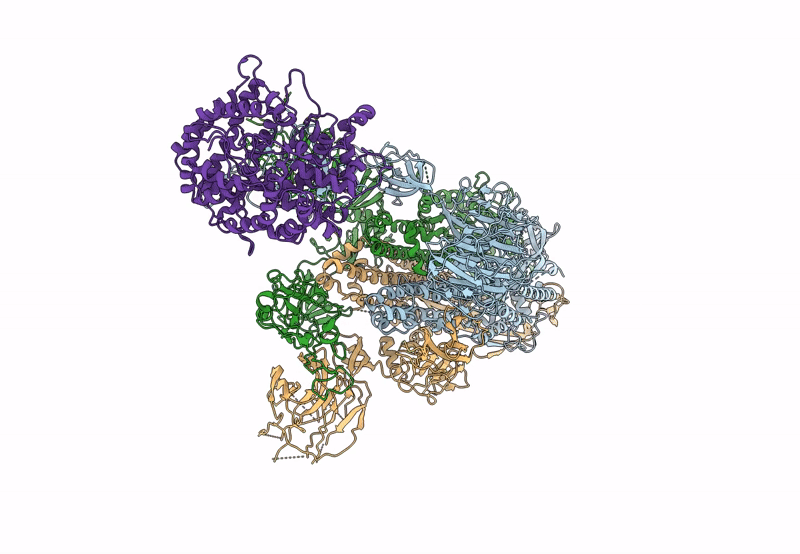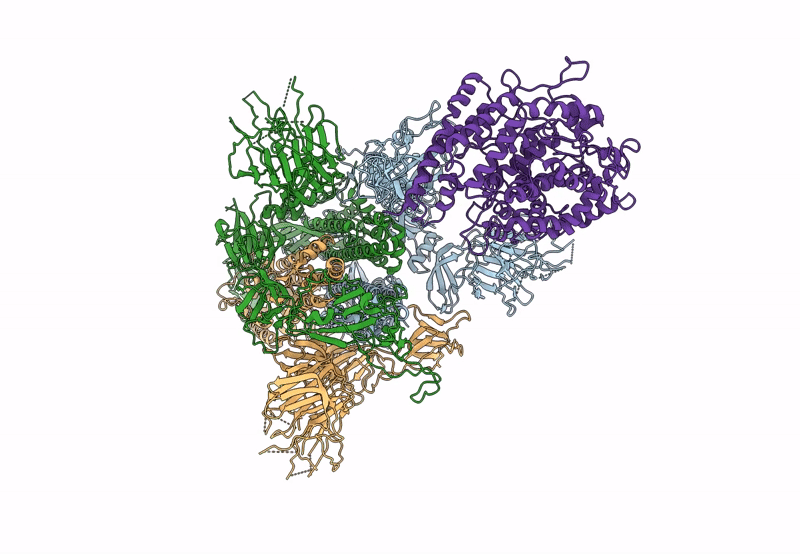
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors
Biological Source:
Source Organism(s):
Severe acute respiratory syndrome coronavirus 2 (Taxon ID: 2697049)
Neogale vison (Taxon ID: 452646)
Neogale vison (Taxon ID: 452646)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.75 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


