Deposition Date
2023-04-26
Release Date
2024-05-01
Last Version Date
2025-05-14
Entry Detail
PDB ID:
8SMO
Keywords:
Title:
Crystal structure of the complex between truncated MLLE domain of PABPC1 and engineered superPAM2 peptide
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
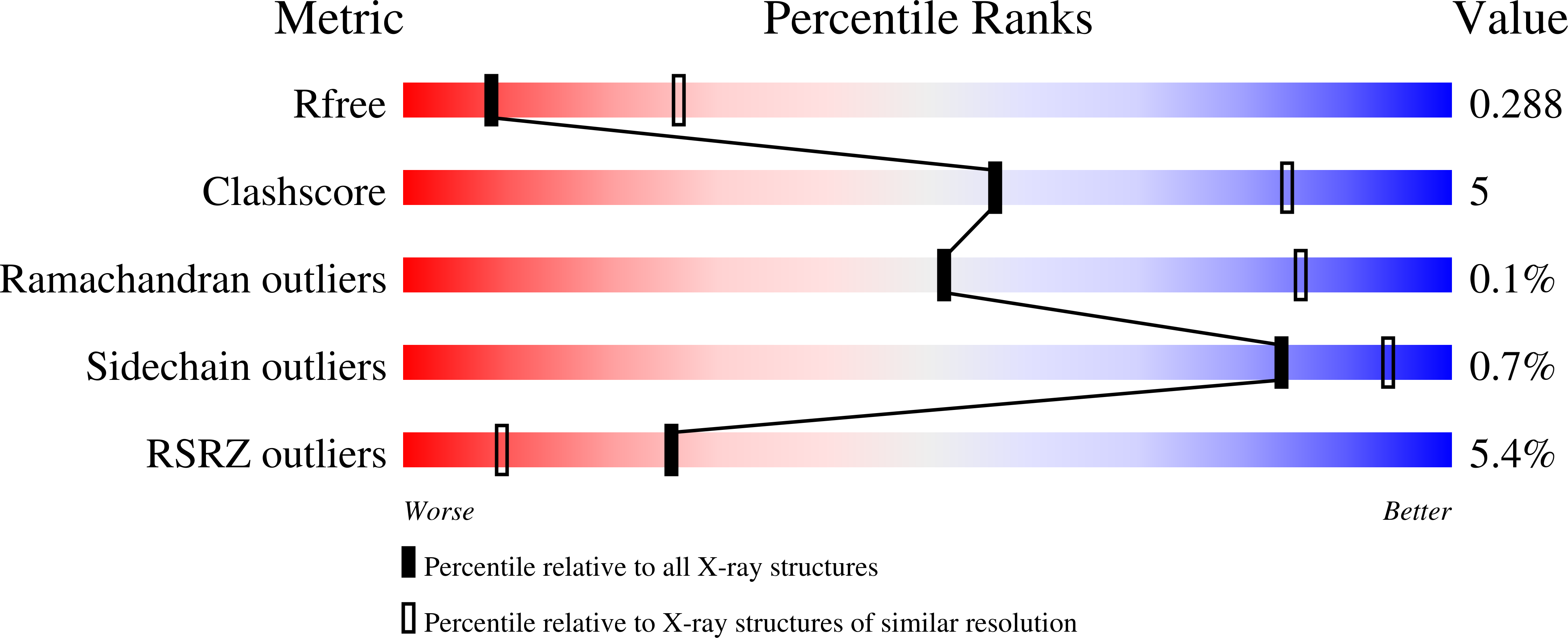
Resolution:
3.00 Å
R-Value Free:
0.29
R-Value Work:
0.25
R-Value Observed:
0.25
Space Group:
P 31