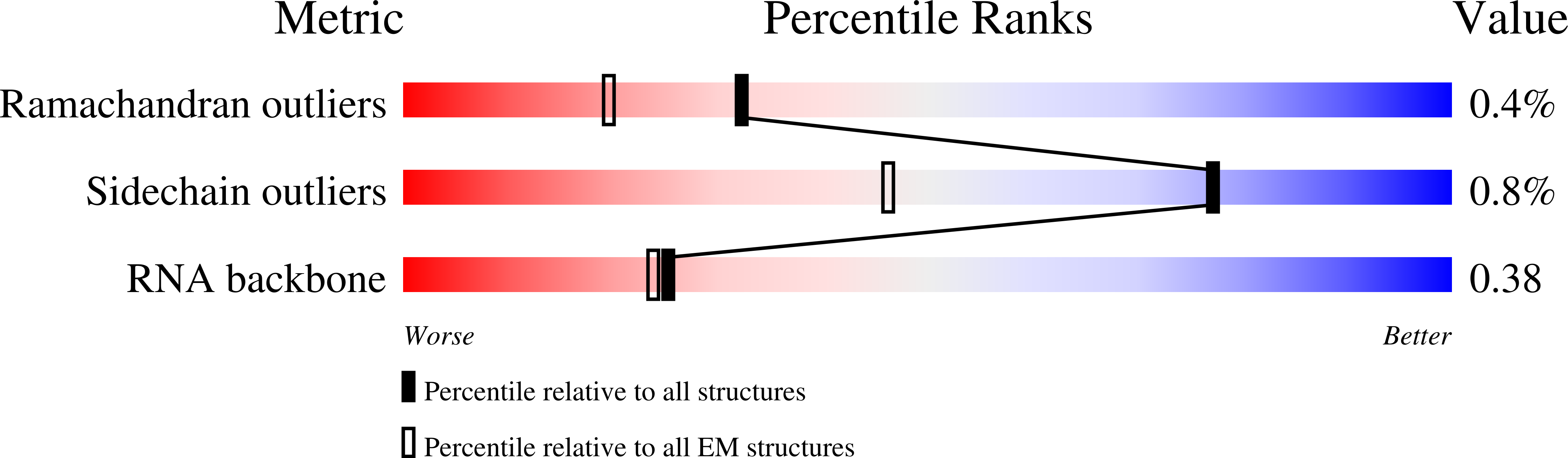
Deposition Date
2024-03-06
Release Date
2024-09-11
Last Version Date
2024-10-23
Entry Detail
PDB ID:
8S8J
Keywords:
Title:
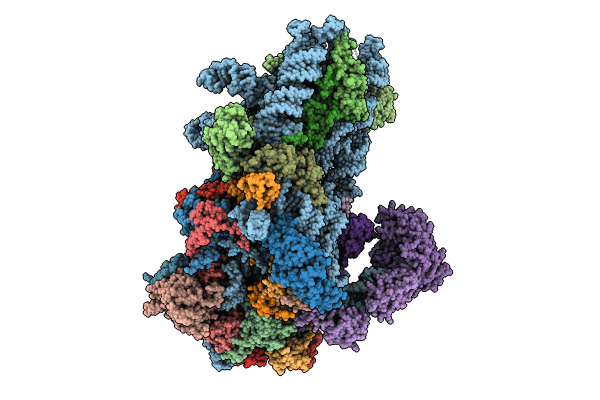
Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5)
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae S288C (Taxon ID: 559292)
Kluyveromyces lactis (Taxon ID: 28985)
Kluyveromyces lactis NRRL Y-1140 (Taxon ID: 284590)
Kluyveromyces lactis (Taxon ID: 28985)
Kluyveromyces lactis NRRL Y-1140 (Taxon ID: 284590)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE