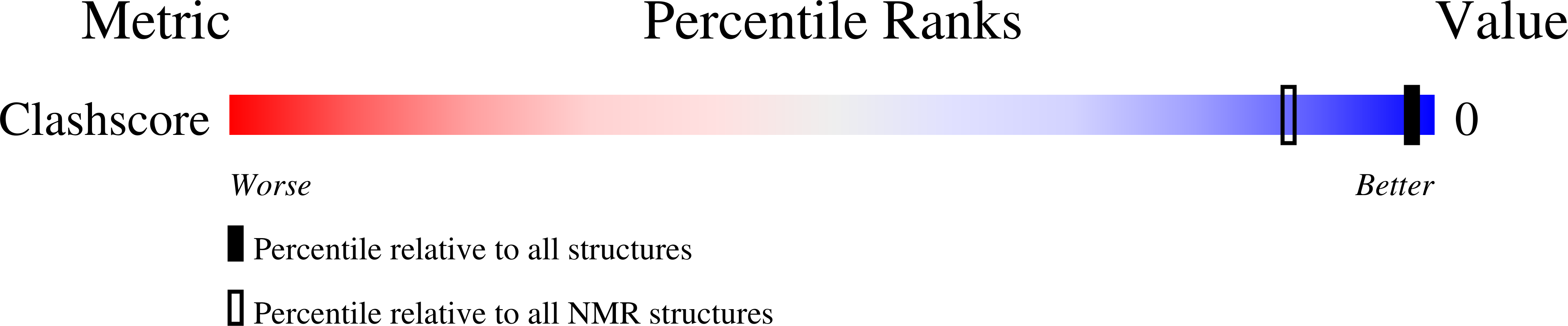Deposition Date
2024-02-21
Release Date
2024-12-18
Last Version Date
2025-01-22
Entry Detail
PDB ID:
8S4N
Keywords:
Title:
An i-motif/B-DNA aptamer with high affinity for influenza A virus
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
10
Selection Criteria:
structures with the lowest energy