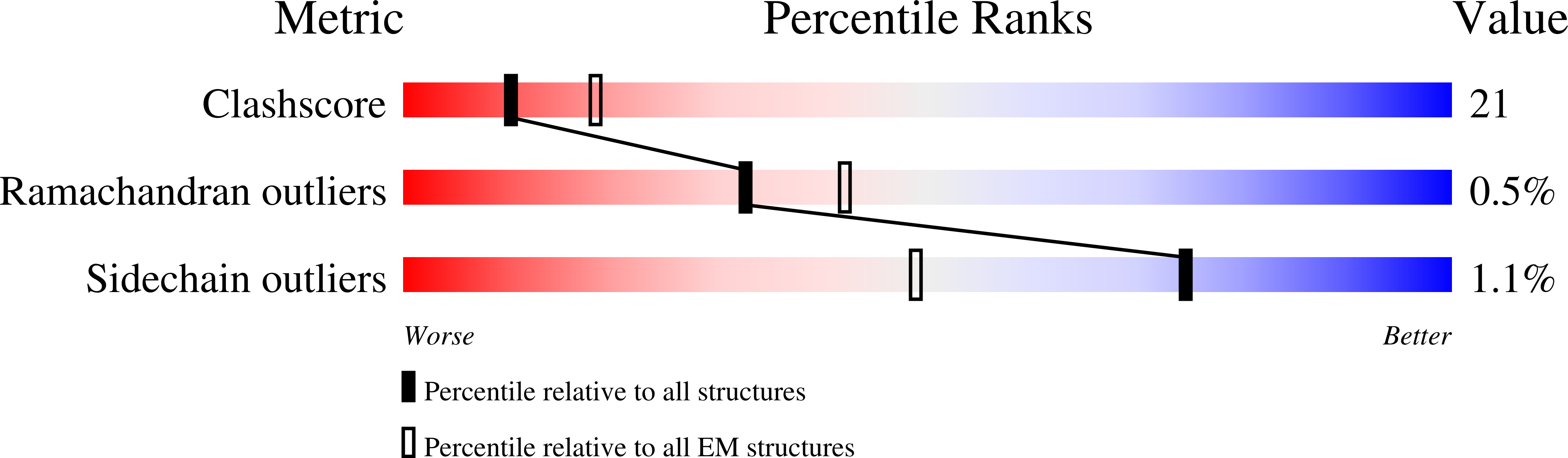
Deposition Date
2024-02-05
Release Date
2024-03-06
Last Version Date
2025-12-24
Entry Detail
PDB ID:
8RX0
Keywords:
Title:
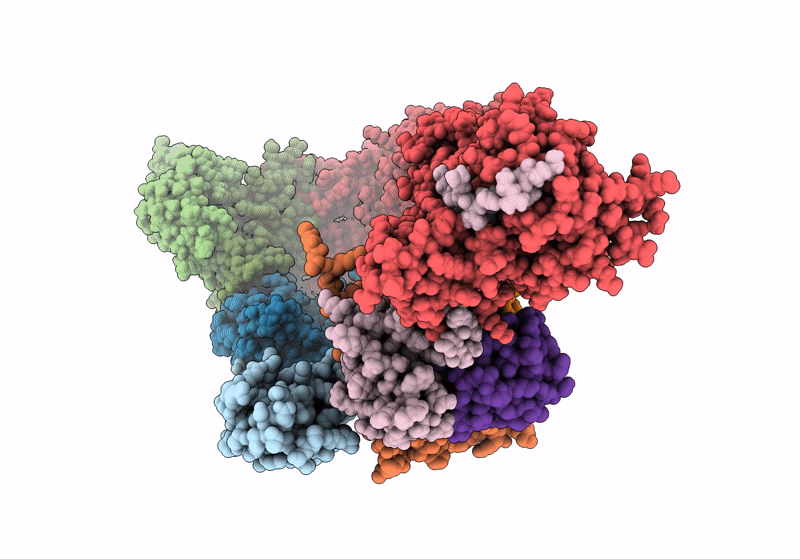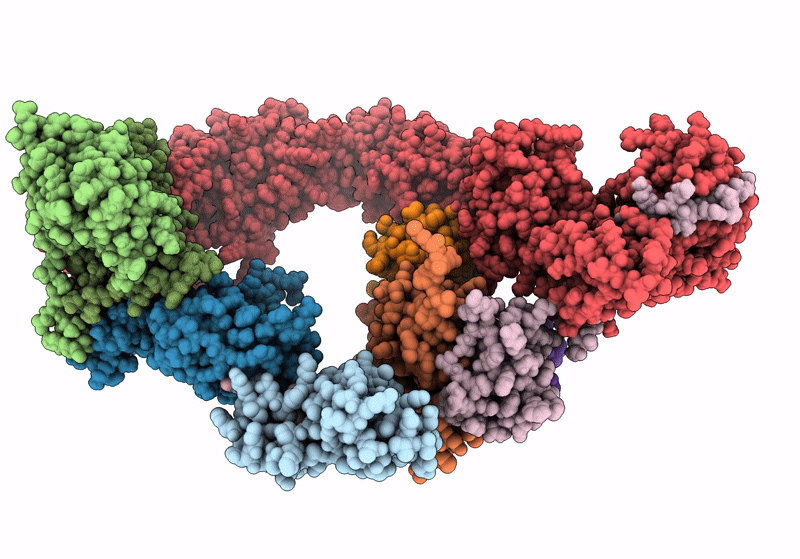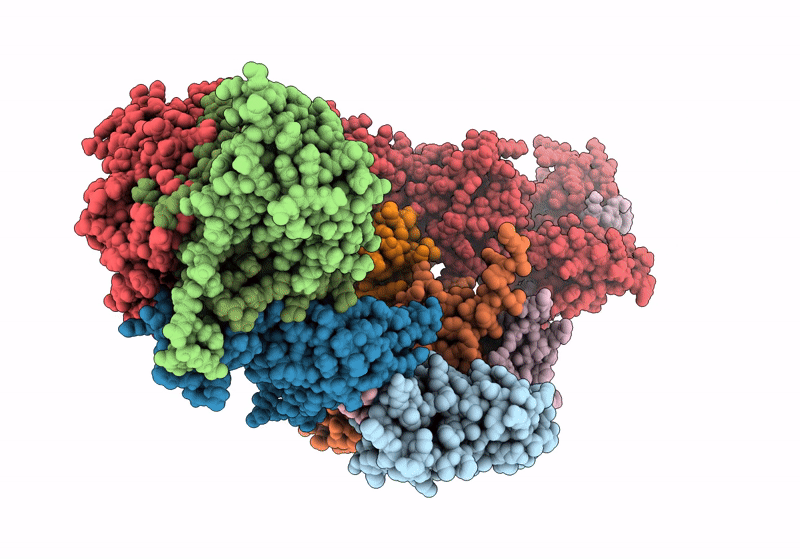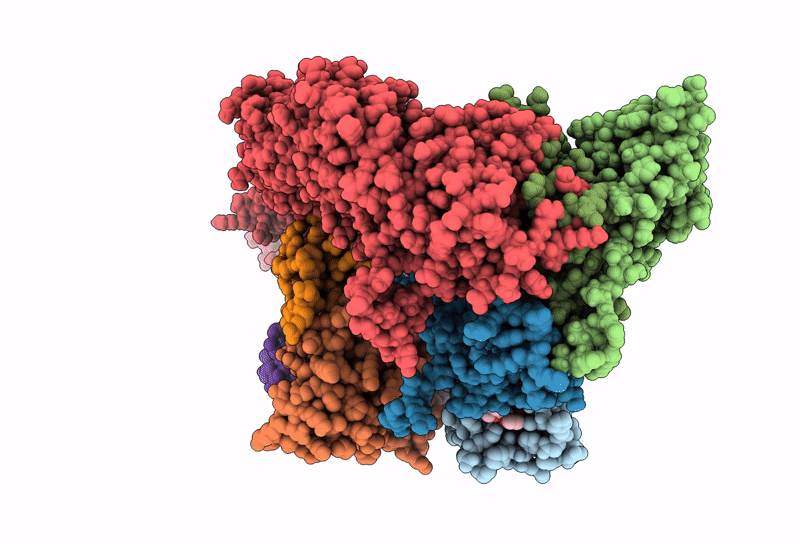
(NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE