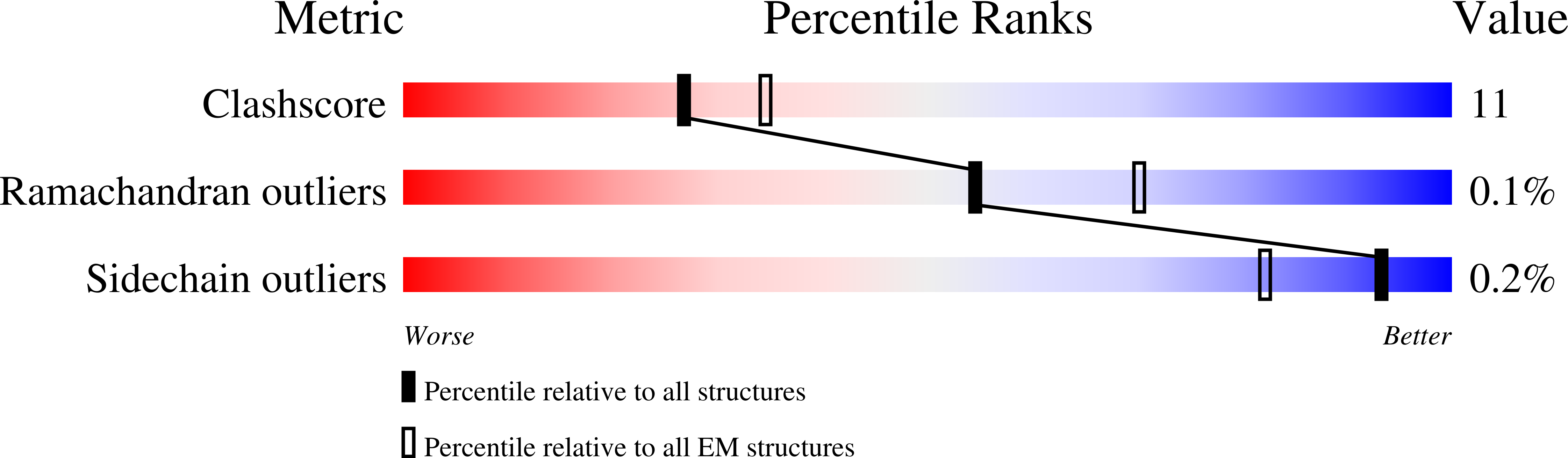
Deposition Date
2024-01-29
Release Date
2024-04-10
Last Version Date
2024-04-24
Entry Detail
PDB ID:
8RTY
Keywords:
Title:
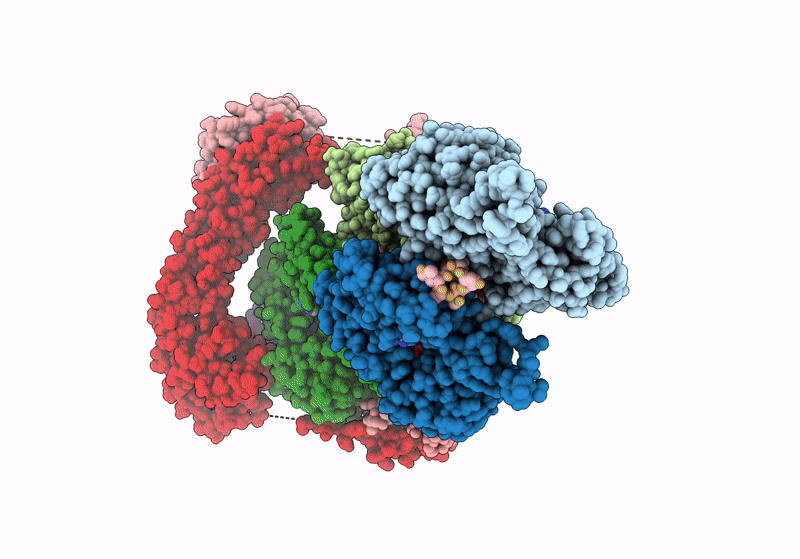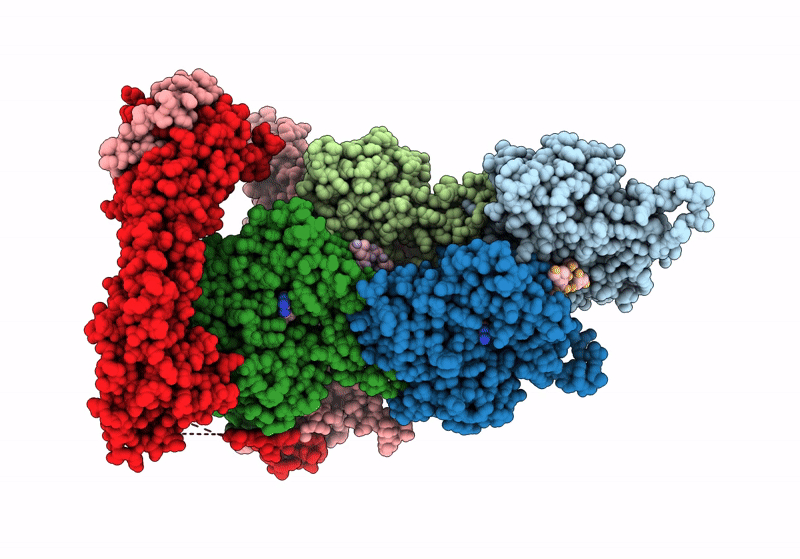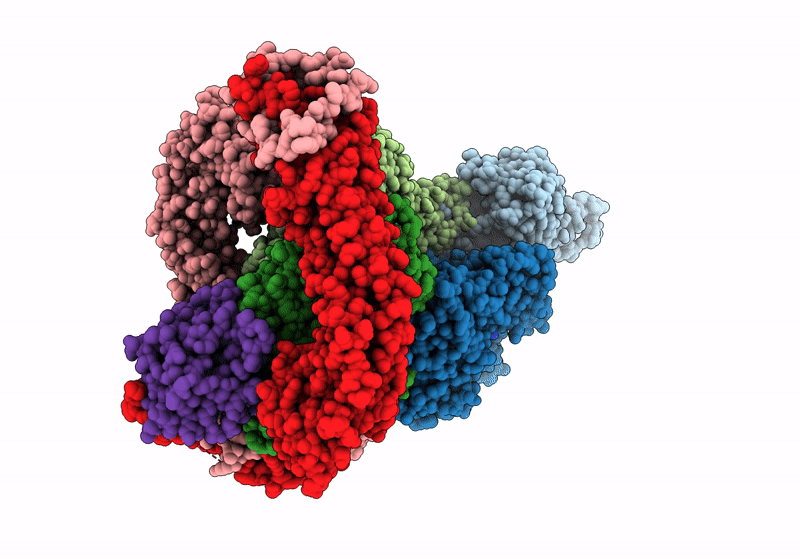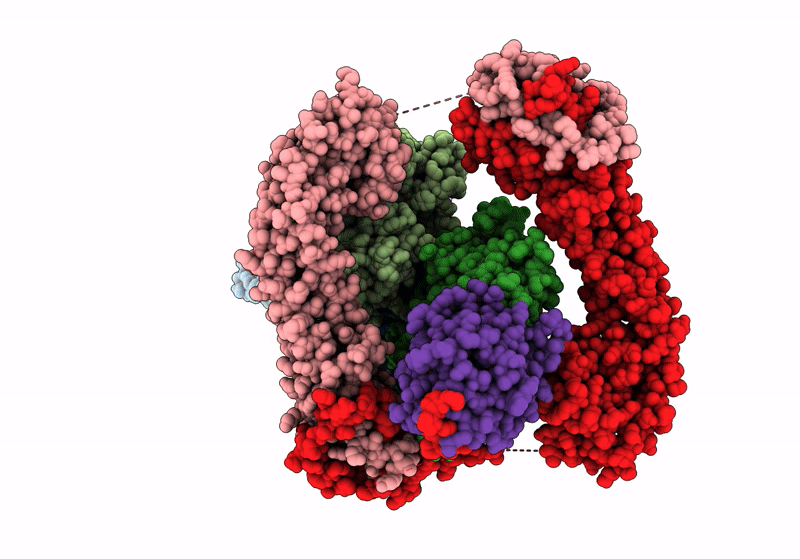
Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Schizosaccharomyces pombe (Taxon ID: 4896)
Amanita phalloides (Taxon ID: 67723)
Schizosaccharomyces pombe (Taxon ID: 4896)
Amanita phalloides (Taxon ID: 67723)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
6.25 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE