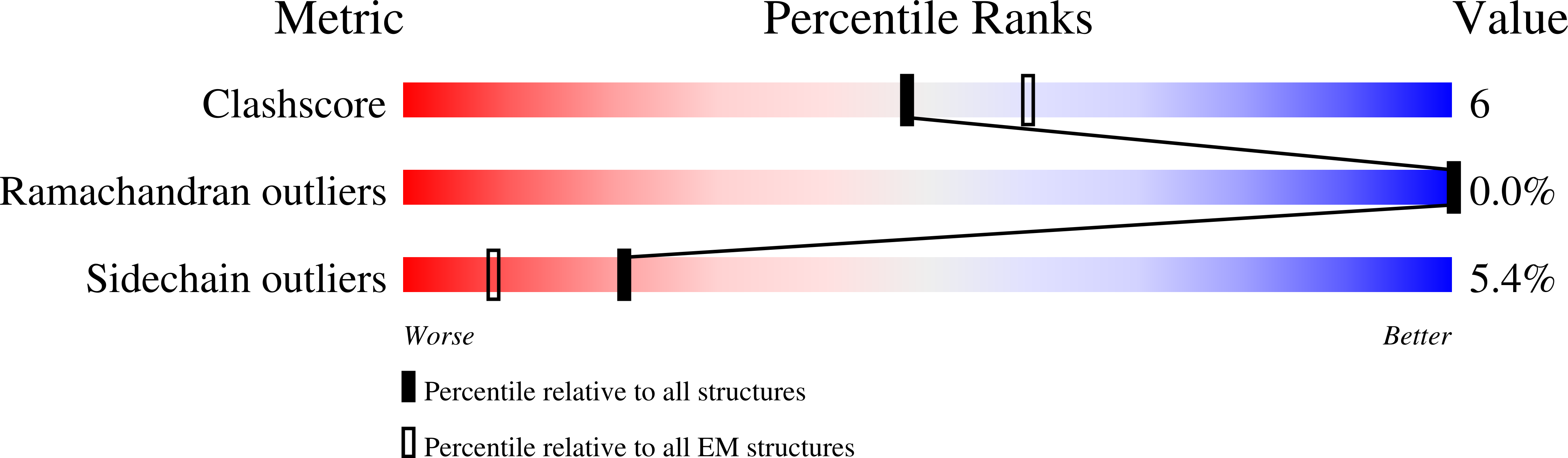
Deposition Date
2024-01-09
Release Date
2024-09-11
Last Version Date
2024-09-11
Entry Detail
PDB ID:
8RN5
Keywords:
Title:
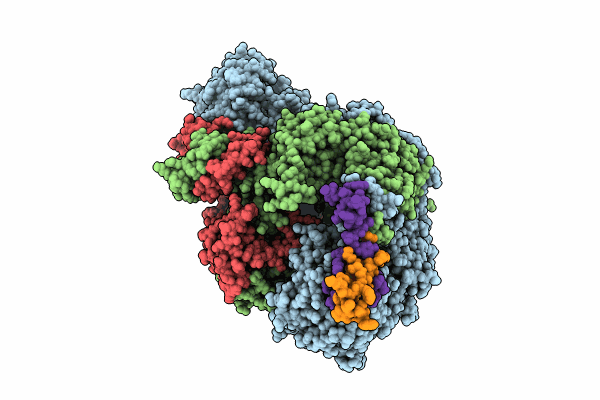
Pseudo-symmetrical influenza B polymerase apo-dimer, ENDO(R) moiety (from "Influenza B polymerase pseudo-symmetrical dimer" | Local refinement)
Biological Source:
Source Organism(s):
Influenza B virus (B/Memphis/13/2003) (Taxon ID: 1601067)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.88 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE