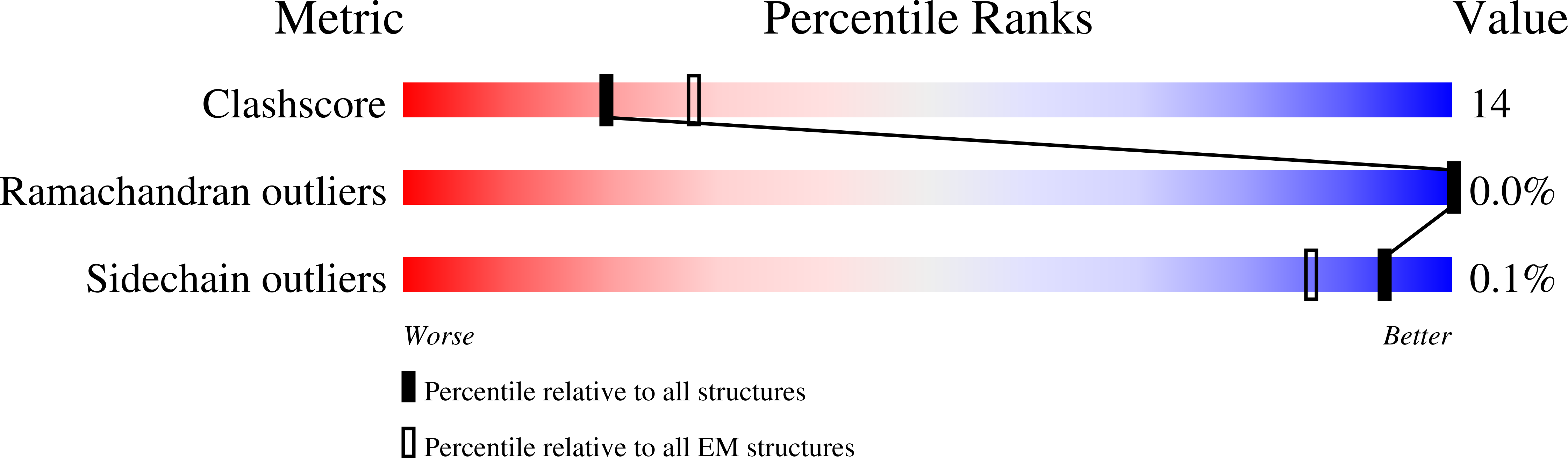
Deposition Date
2023-10-17
Release Date
2024-04-24
Last Version Date
2024-07-31
Entry Detail
PDB ID:
8QV3
Keywords:
Title:
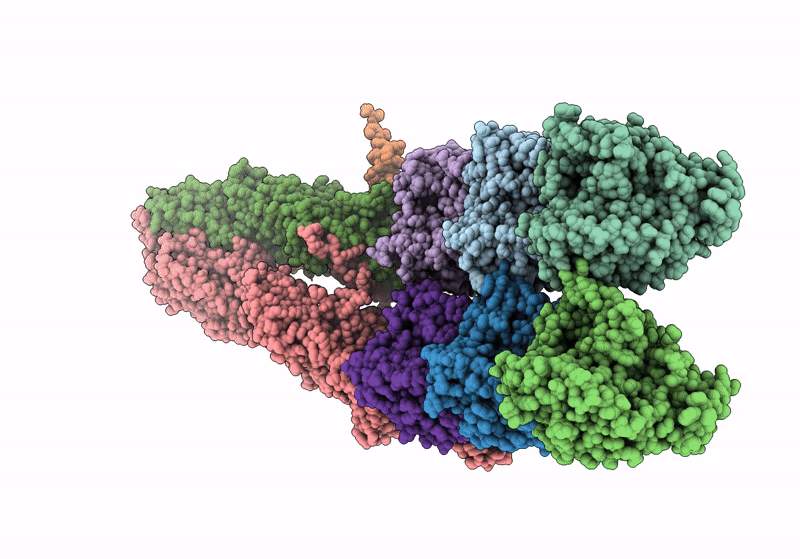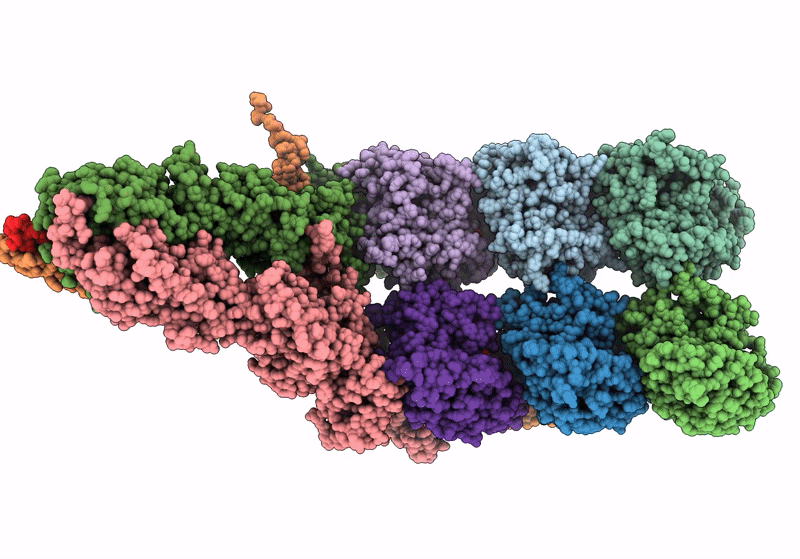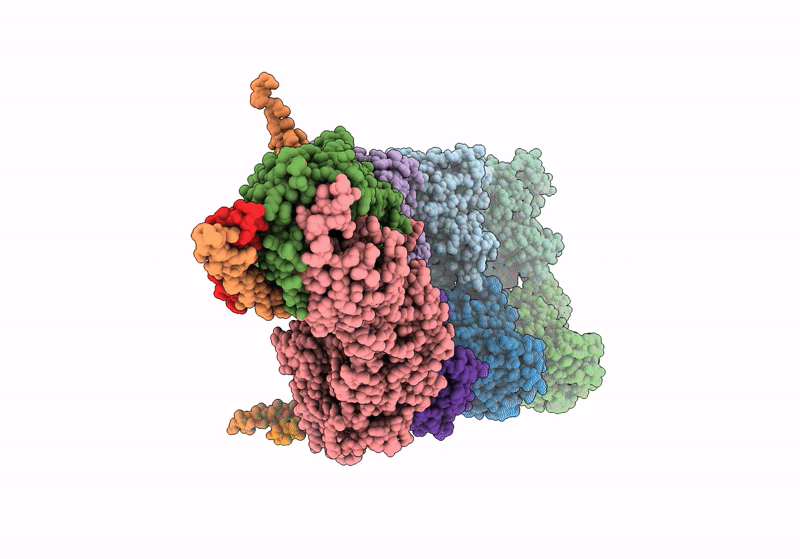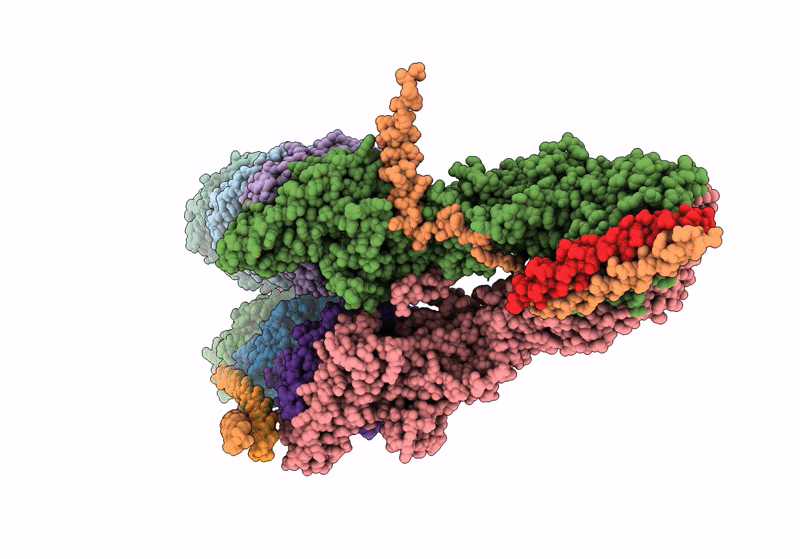
Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Method Details:
Experimental Method:
Resolution:
8.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SUBTOMOGRAM AVERAGING