Deposition Date
2023-10-13
Release Date
2024-06-26
Last Version Date
2024-07-17
Entry Detail
PDB ID:
8QU1
Keywords:
Title:
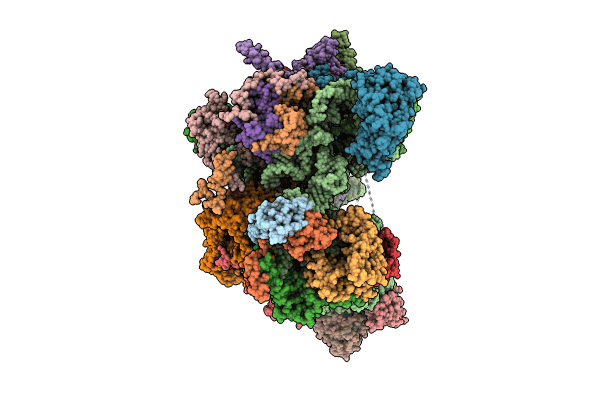
mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 1
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
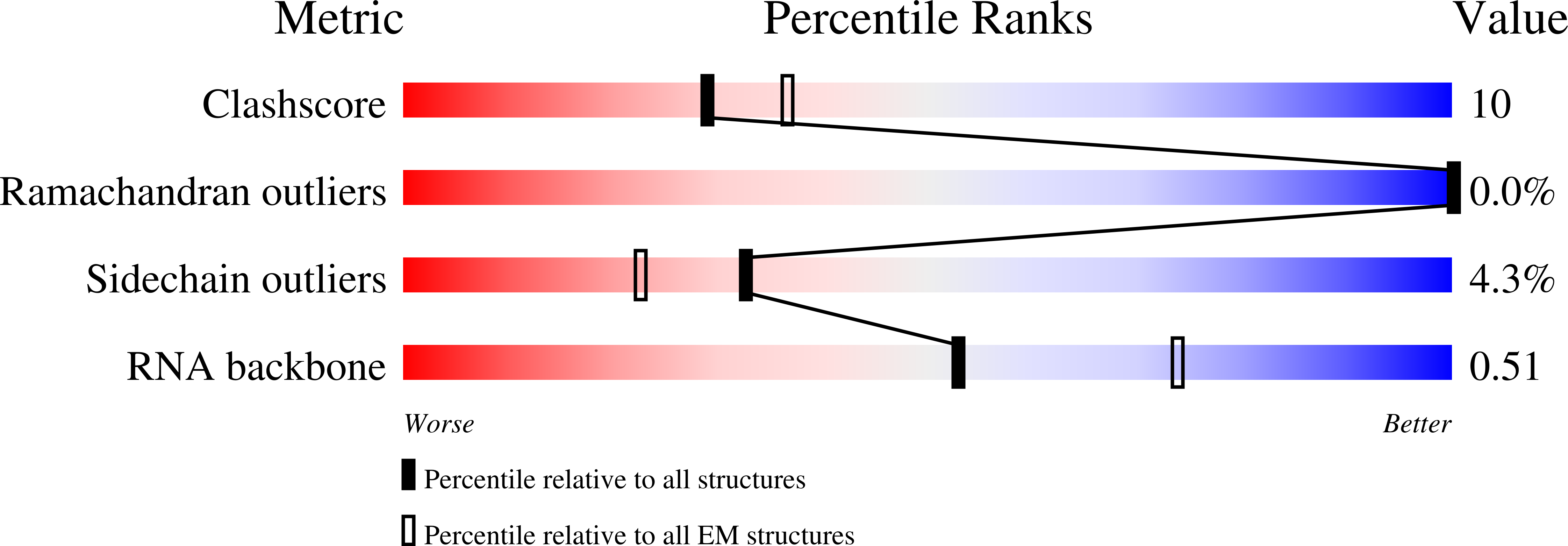
Resolution:
2.74 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE