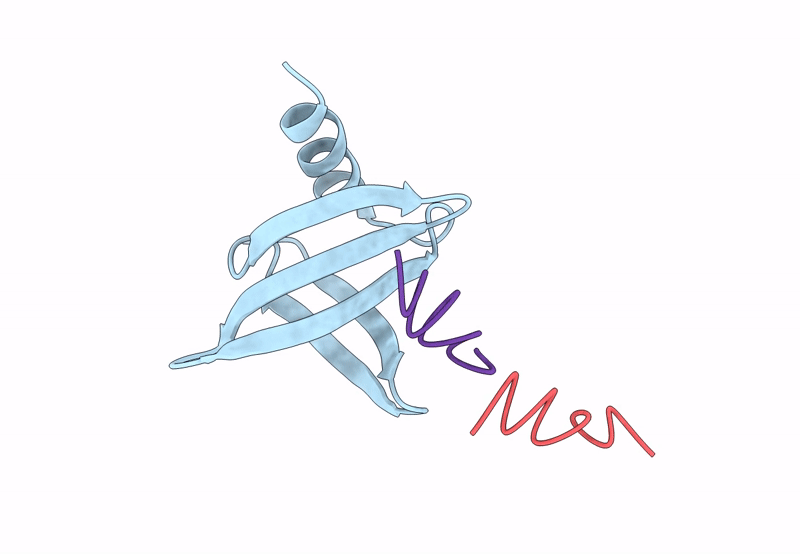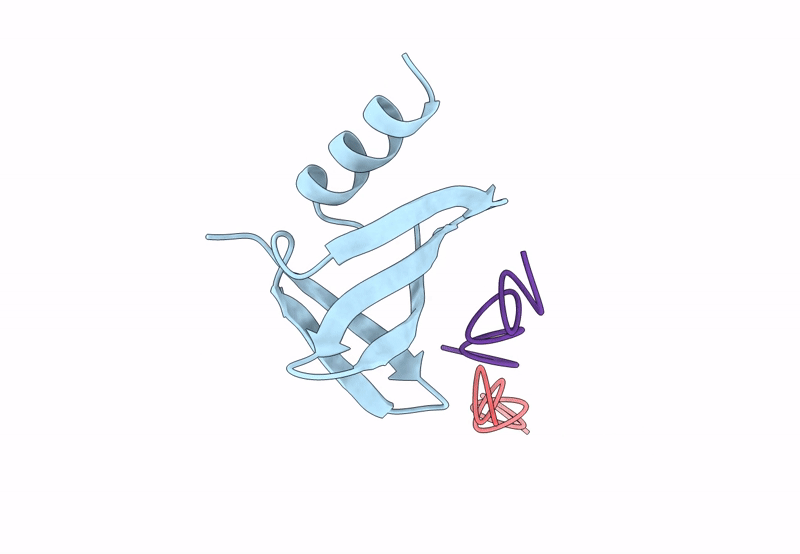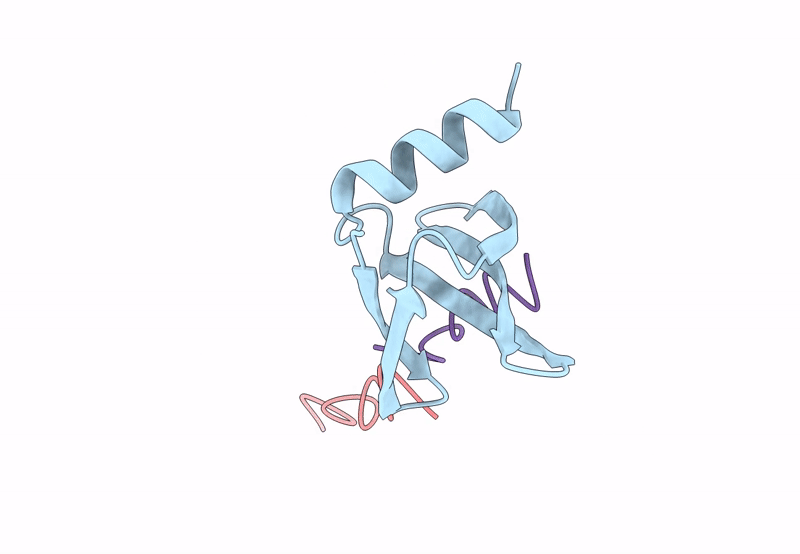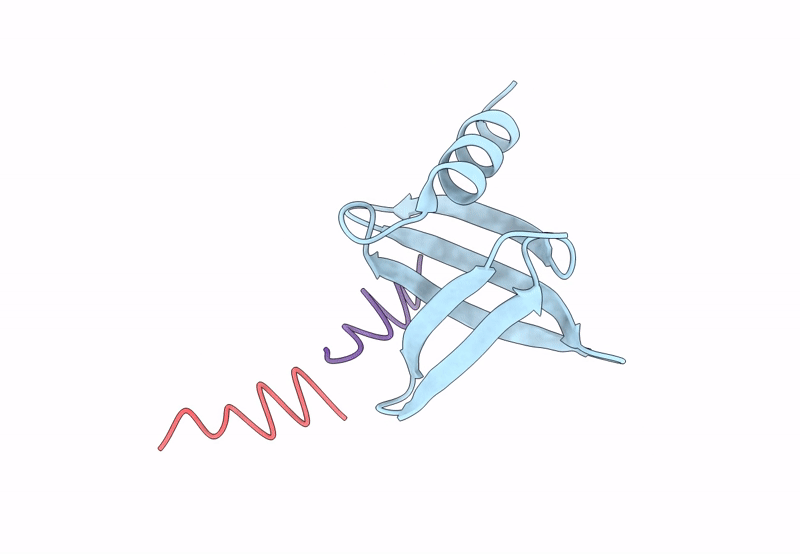
Deposition Date
2023-10-01
Release Date
2023-11-08
Last Version Date
2025-02-26
Entry Detail
PDB ID:
8QPC
Keywords:
Title:
18mer DNA mimic Foldamer with an Aromatic linker in complex with Sac7d V26A/M29A protein
Biological Source:
Source Organism(s):
Sulfolobus acidocaldarius (Taxon ID: 2285)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.24 Å
R-Value Free:
0.34
R-Value Work:
0.33
R-Value Observed:
0.33
Space Group:
P 64 2 2


