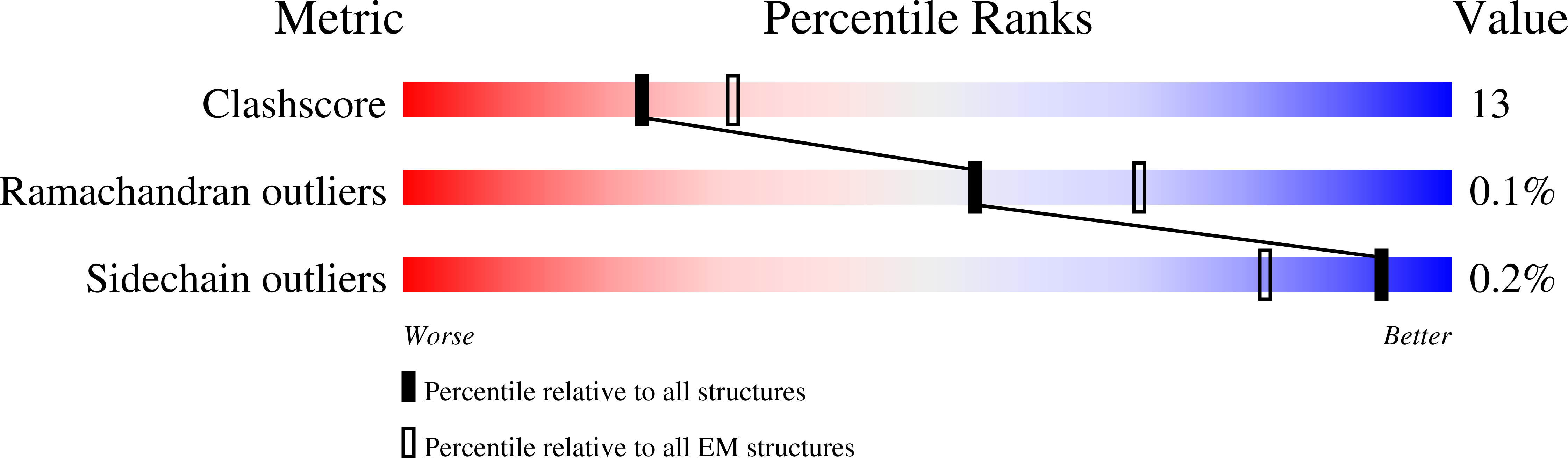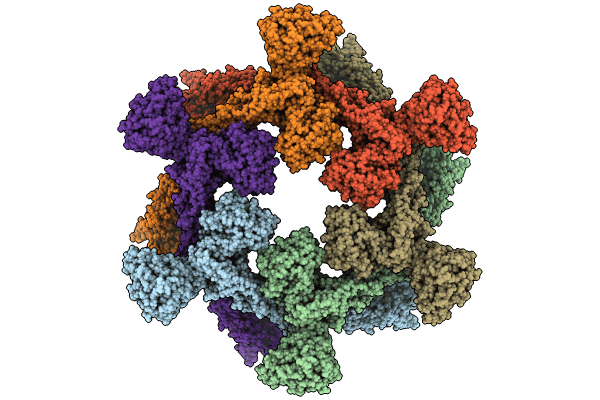
Deposition Date
2023-09-29
Release Date
2024-02-21
Last Version Date
2024-11-13
Entry Detail
PDB ID:
8QP0
Keywords:
Title:
A hexamer pore in the S-layer of Sulfolobus acidocaldarius formed by SlaA protein
Biological Source:
Source Organism(s):
Sulfolobus acidocaldarius DSM 639 (Taxon ID: 330779)
Method Details:
Experimental Method:
Resolution:
11.20 Å
Aggregation State:
3D ARRAY
Reconstruction Method:
SUBTOMOGRAM AVERAGING