Deposition Date
2023-08-30
Release Date
2024-06-26
Last Version Date
2024-07-31
Entry Detail
PDB ID:
8QE4
Keywords:
Title:
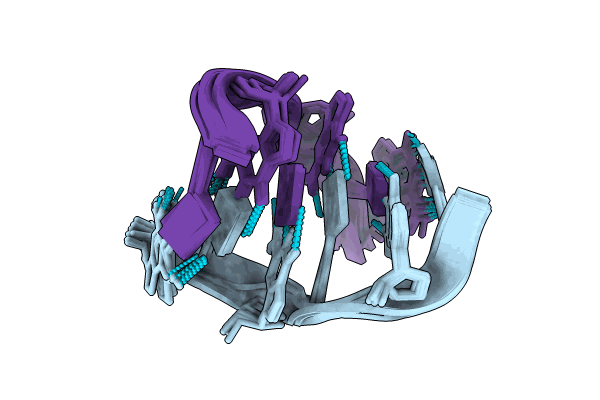
Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Conformers Calculated:
10
Conformers Submitted:
10
Selection Criteria:
all calculated structures submitted