Deposition Date
2023-08-11
Release Date
2023-09-20
Last Version Date
2024-10-23
Entry Detail
PDB ID:
8Q6J
Keywords:
Title:
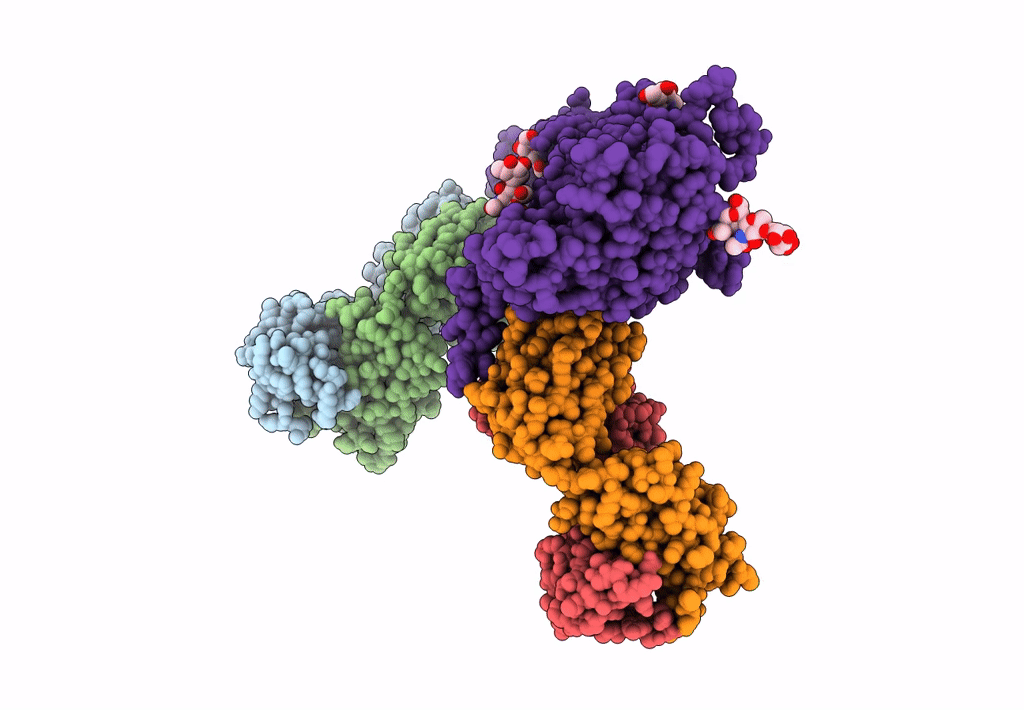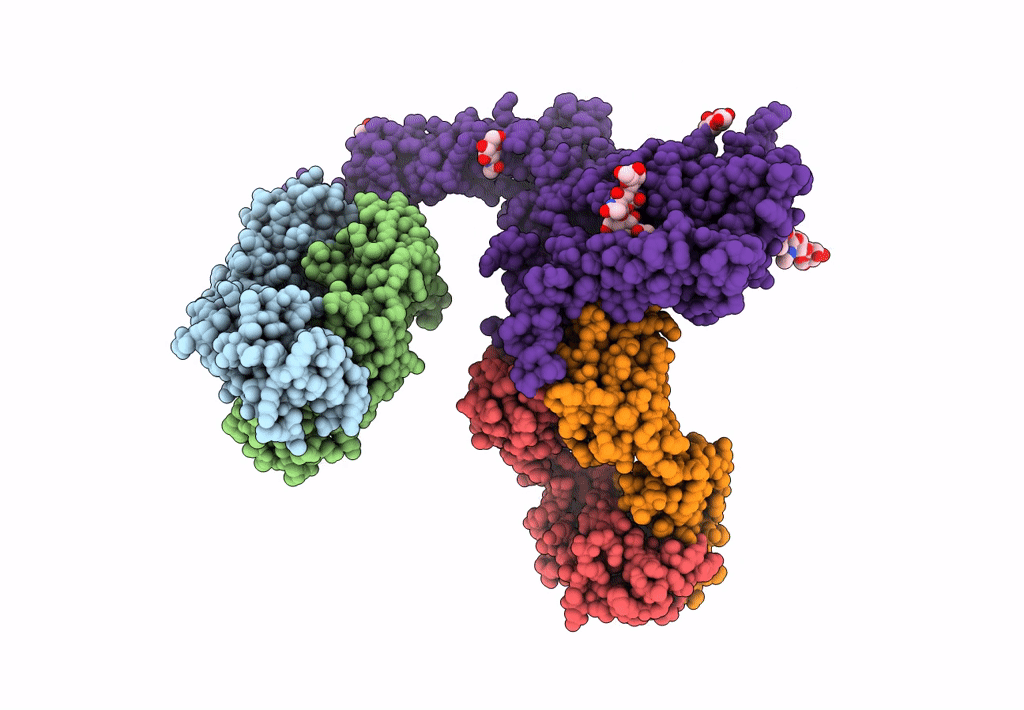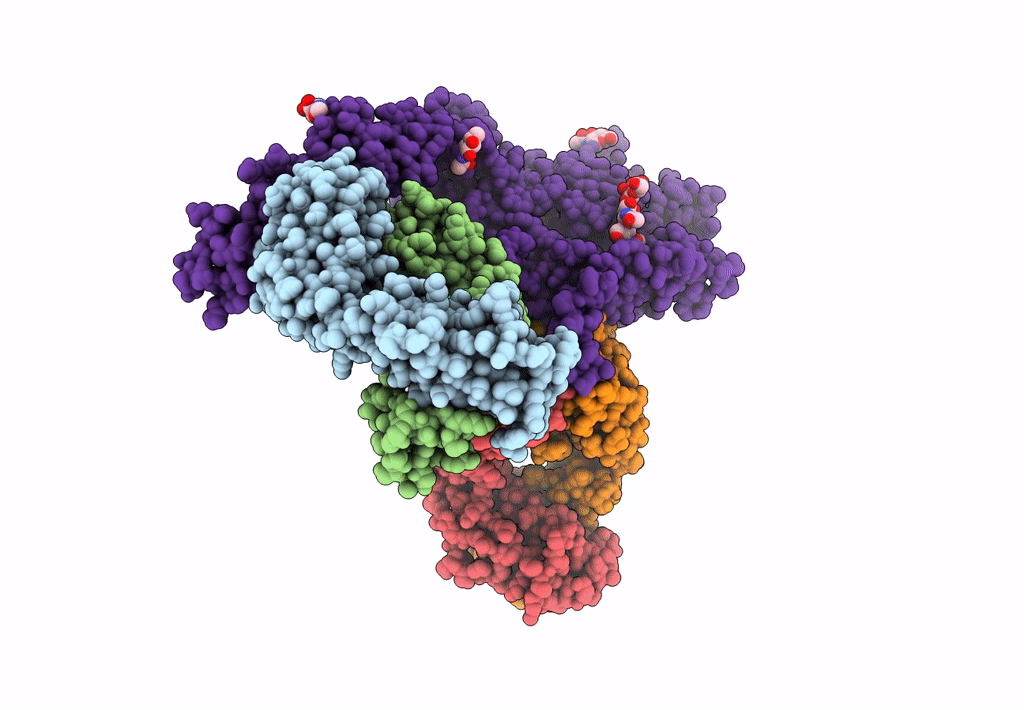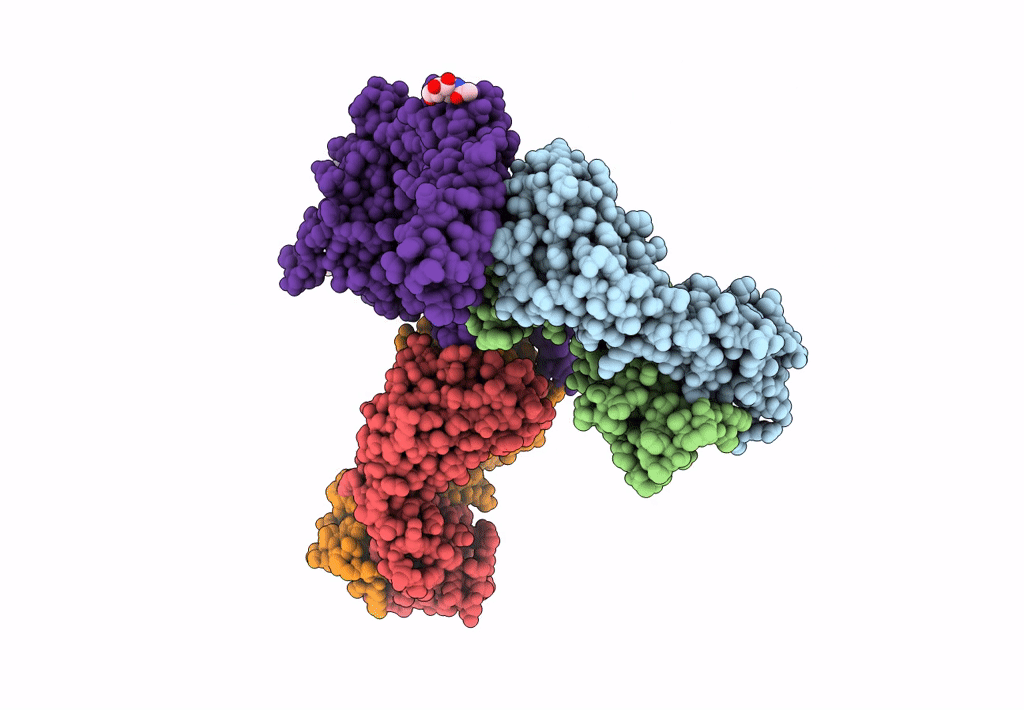
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Resolution:
3.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


