Deposition Date
2023-07-20
Release Date
2025-02-05
Last Version Date
2025-04-23
Entry Detail
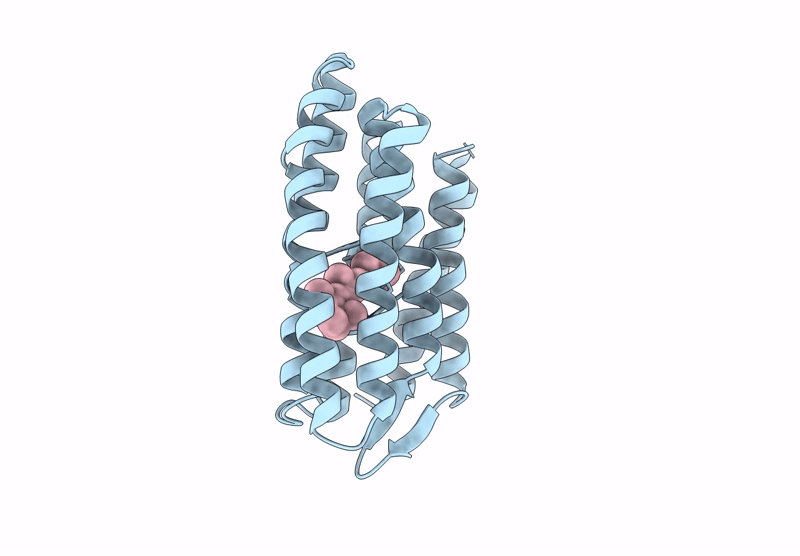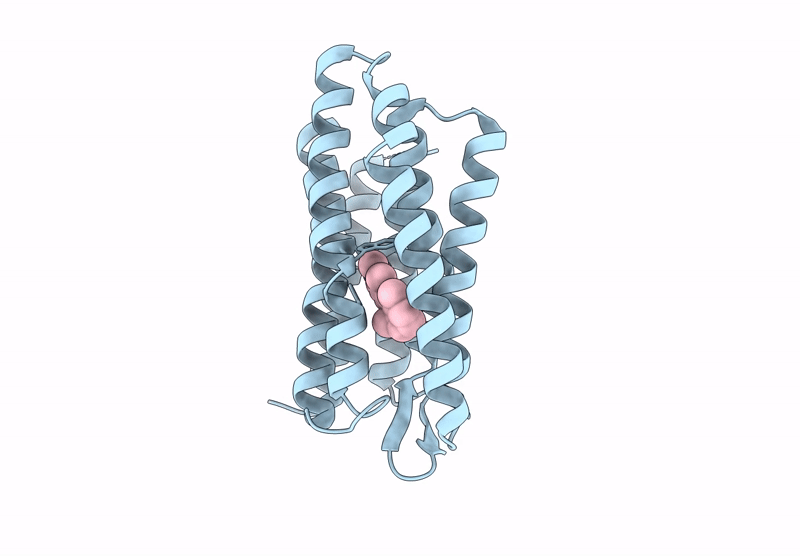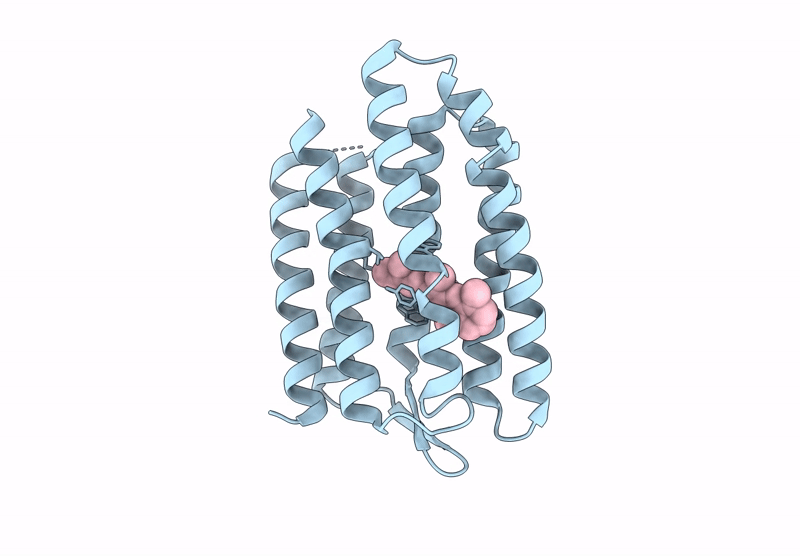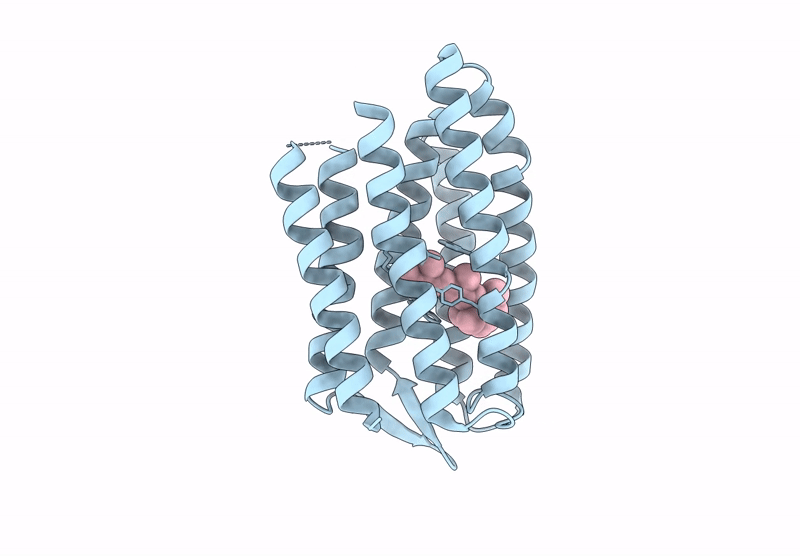
PDB ID:
8PWJ
Keywords:
Title:
Light structure of sensory rhodopsin-II solved by serial millisecond crystallography. 30-60 milliseconds time-bin
Biological Source:
Source Organism(s):
Natronomonas pharaonis (Taxon ID: 2257)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.14 Å
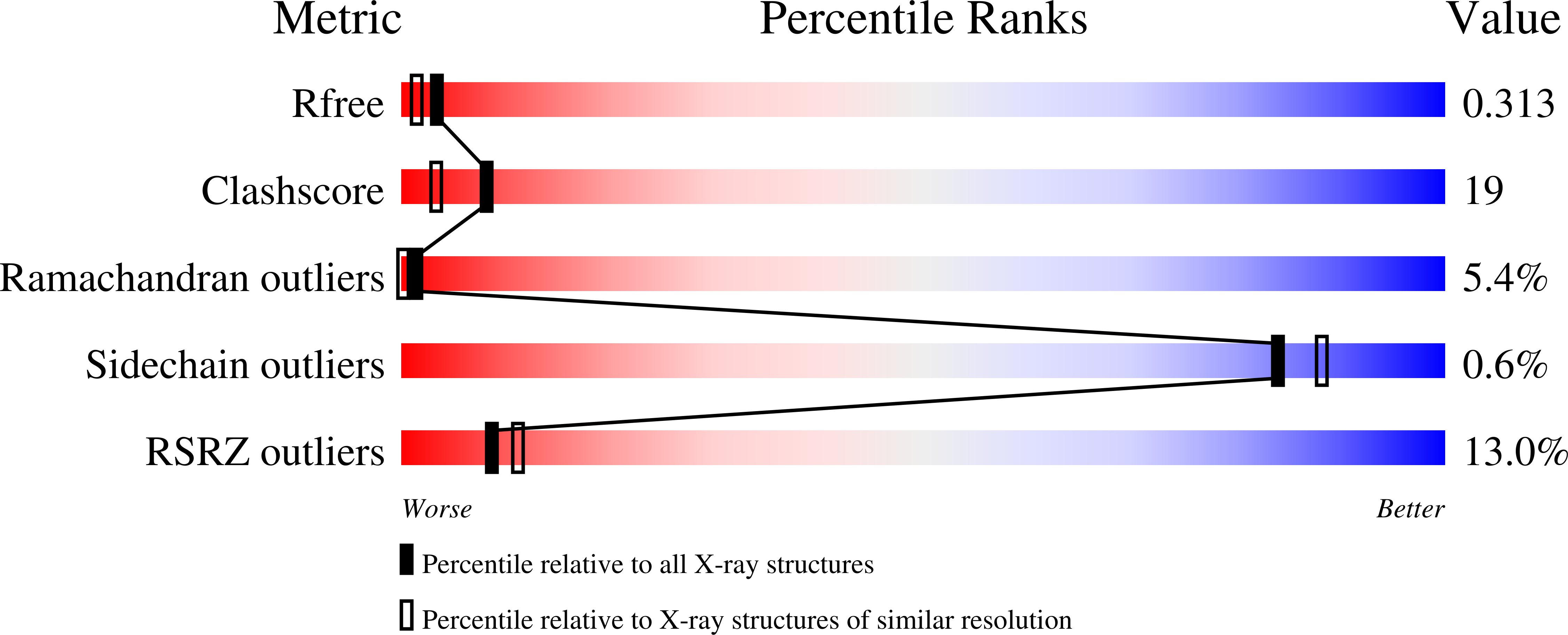
R-Value Free:
0.31
R-Value Work:
0.24
R-Value Observed:
0.25
Space Group:
C 2 2 21