Deposition Date
2023-06-12
Release Date
2023-12-06
Last Version Date
2024-03-20
Entry Detail
PDB ID:
8PDQ
Keywords:
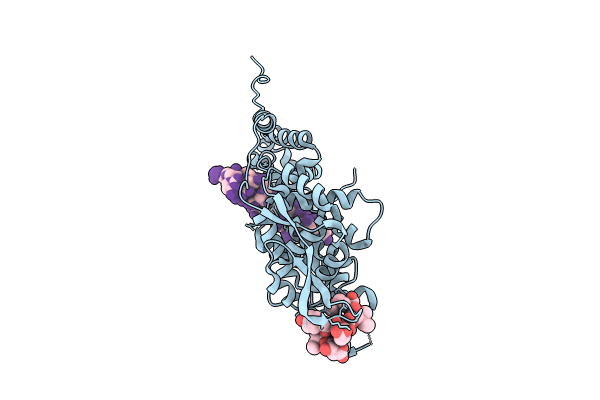
Title:
11-mer ring of HMPV N-RNA bound to the C-terminal region of P
Biological Source:
Source Organism(s):
Human metapneumovirus (strain CAN97-83) (Taxon ID: 694067)
Escherichia coli (Taxon ID: 562)
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
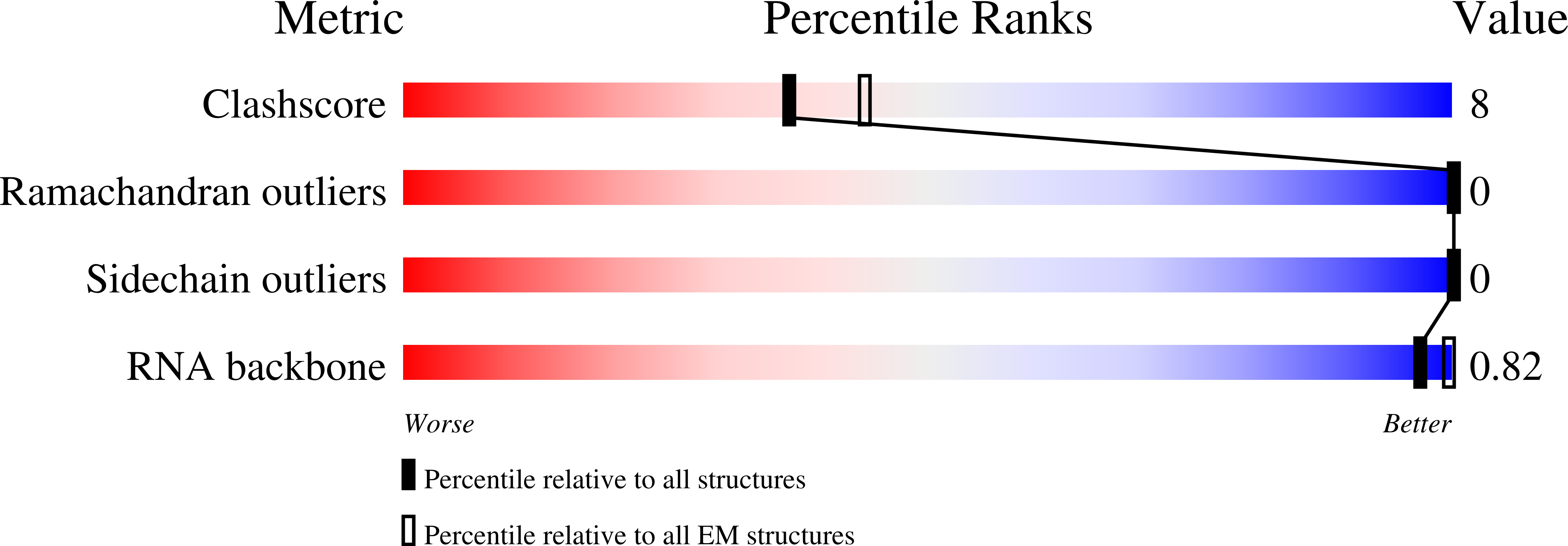
Resolution:
3.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE