Deposition Date
2023-06-04
Release Date
2024-06-12
Last Version Date
2026-01-07
Entry Detail
PDB ID:
8JM0
Keywords:
Title:
Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A from Prunus communis complexed with 2,2-dimethyl-4H-benzo[d][1,3]dioxine-6-carbaldehyde from the cyanohydrin cleavage
Biological Source:
Source Organism(s):
Prunus dulcis (Taxon ID: 3755)
Expression System(s):
Method Details:
Experimental Method:
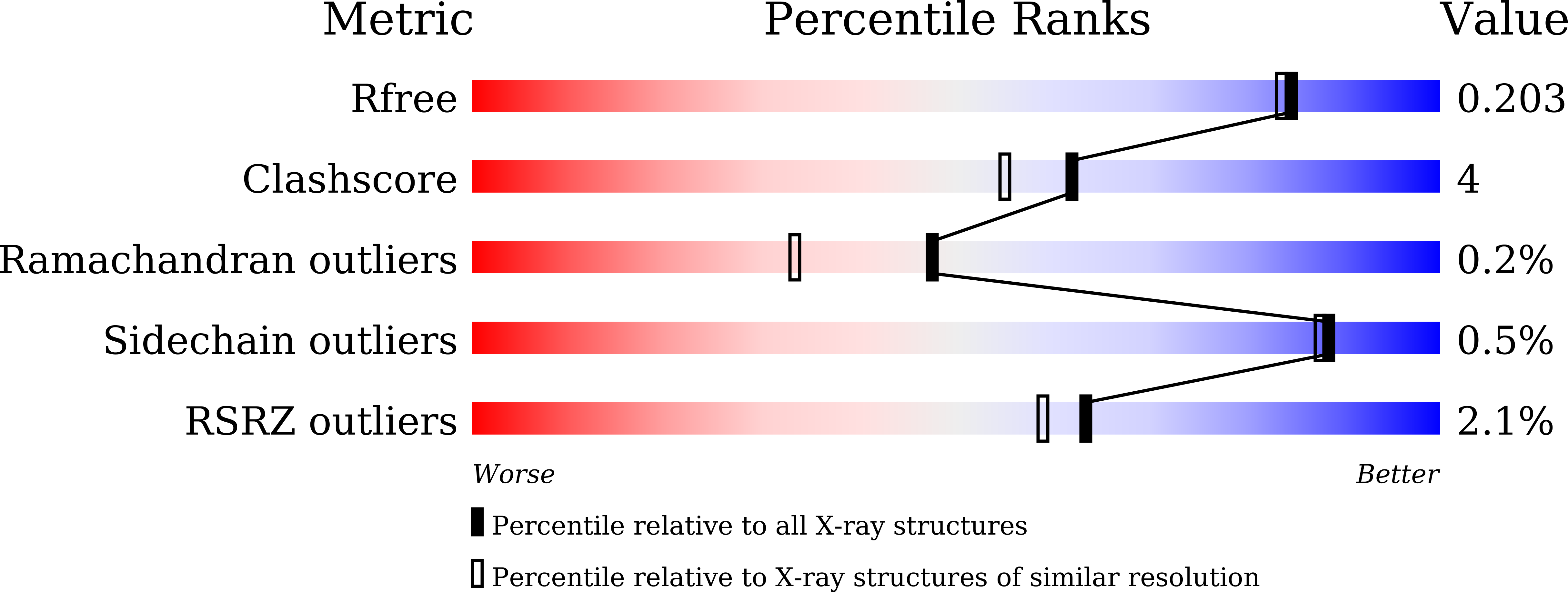
Resolution:
1.79 Å
R-Value Free:
0.19
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 21 21 21