Deposition Date
2022-11-28
Release Date
2023-08-16
Last Version Date
2024-05-22
Entry Detail
PDB ID:
8HKX
Keywords:
Title:
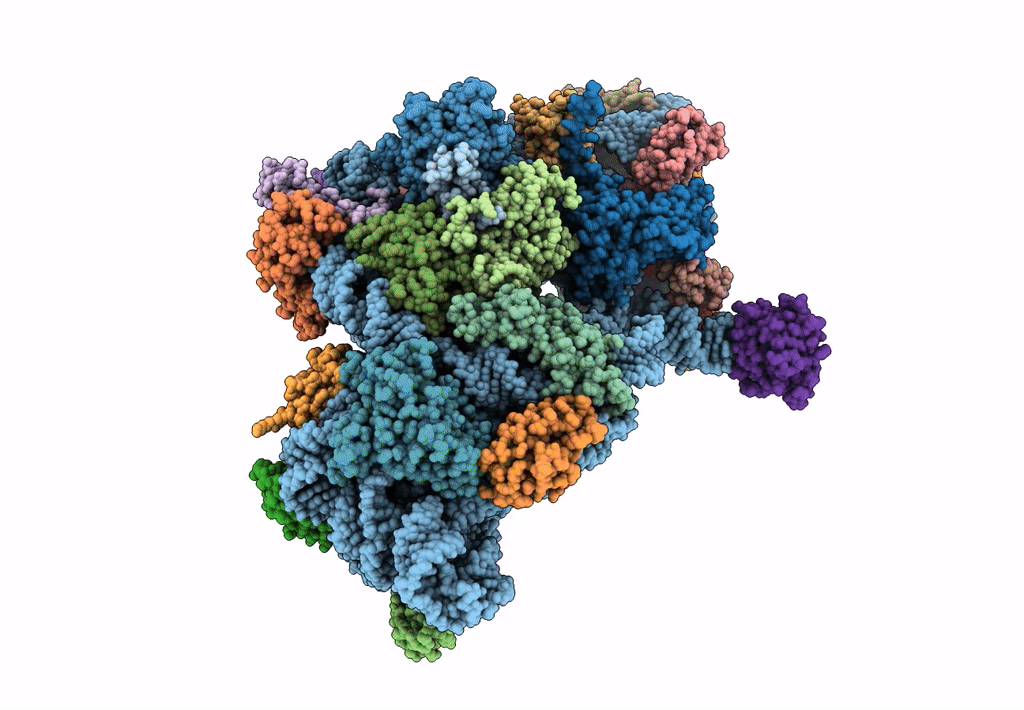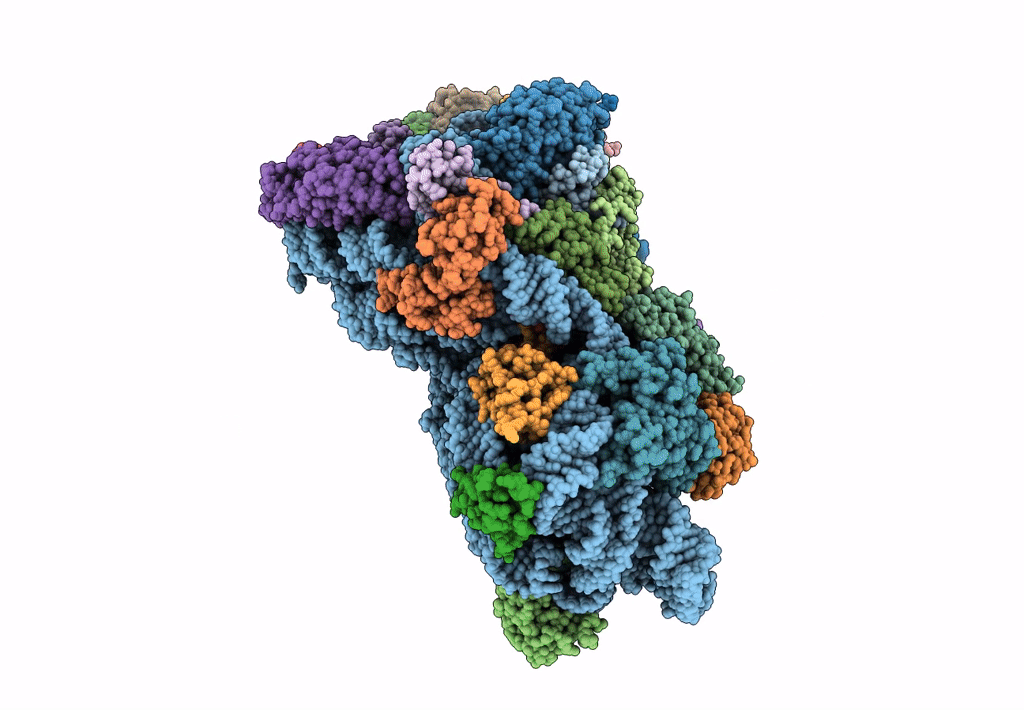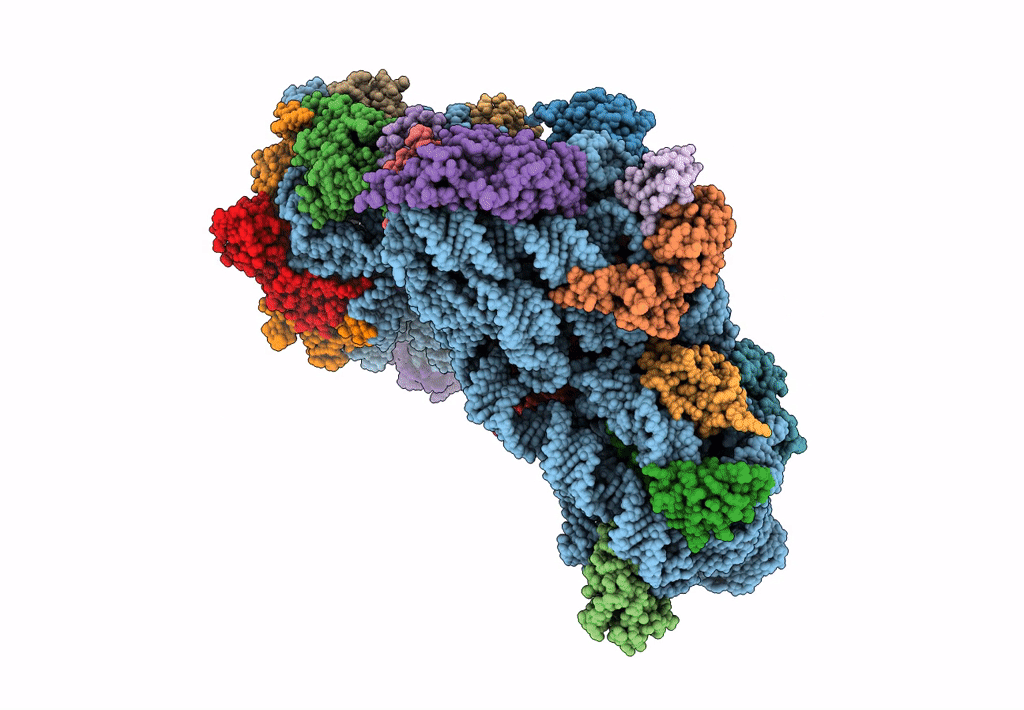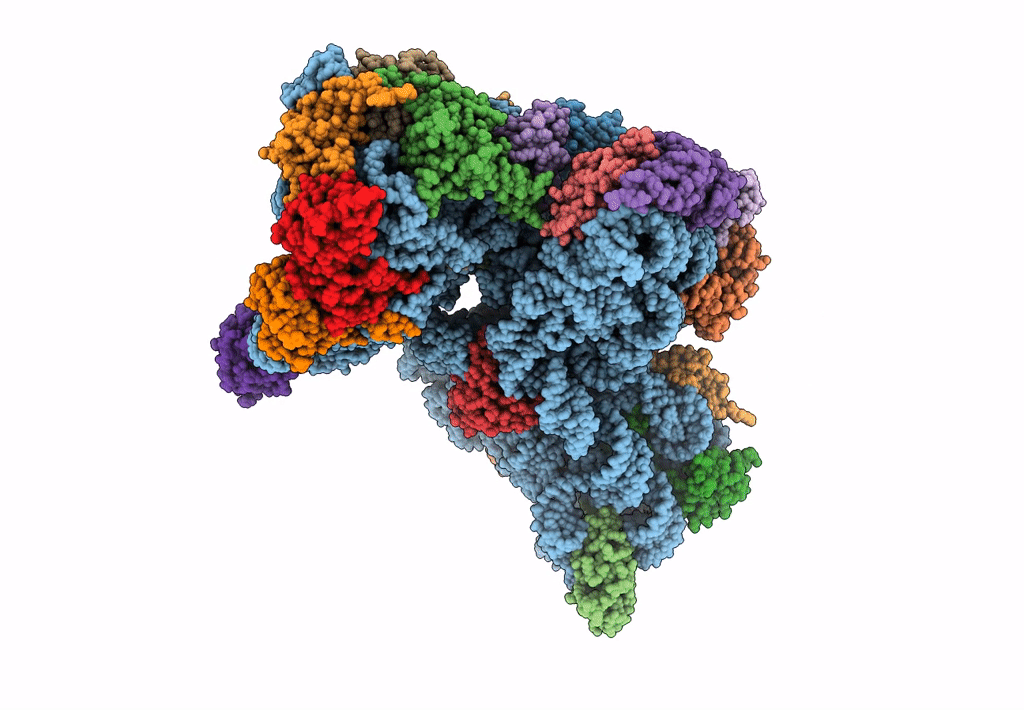
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome
Biological Source:
Source Organism:
Sulfolobus acidocaldarius DSM 639 (Taxon ID: 330779)
Method Details:
Experimental Method:
Resolution:
3.14 Å
Aggregation State:
2D ARRAY
Reconstruction Method:
SINGLE PARTICLE


