Deposition Date
2022-09-11
Release Date
2023-02-15
Last Version Date
2023-11-29
Entry Detail
PDB ID:
8GUE
Keywords:
Title:
Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238
Biological Source:
Source Organism(s):
Streptomyces venezuelae (Taxon ID: 54571)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.90 Å
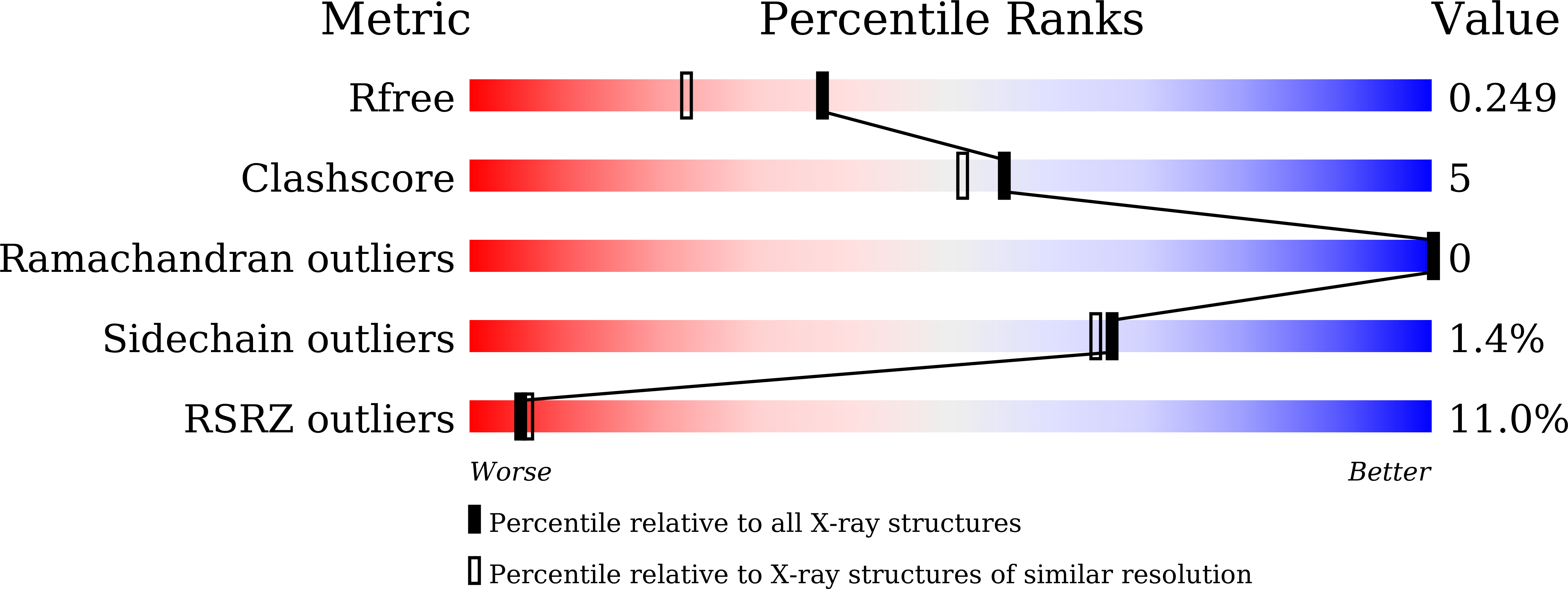
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 21