Deposition Date
2022-08-28
Release Date
2023-08-30
Last Version Date
2024-10-16
Entry Detail
PDB ID:
8GQ1
Keywords:
Title:
HyHEL10 Fab complexed with hen egg lysozyme carrying arginine cluster in framework region of light chain.
Biological Source:
Source Organism:
Mus musculus (Taxon ID: 10090)
Gallus gallus (Taxon ID: 9031)
Gallus gallus (Taxon ID: 9031)
Host Organism:
Method Details:
Experimental Method:
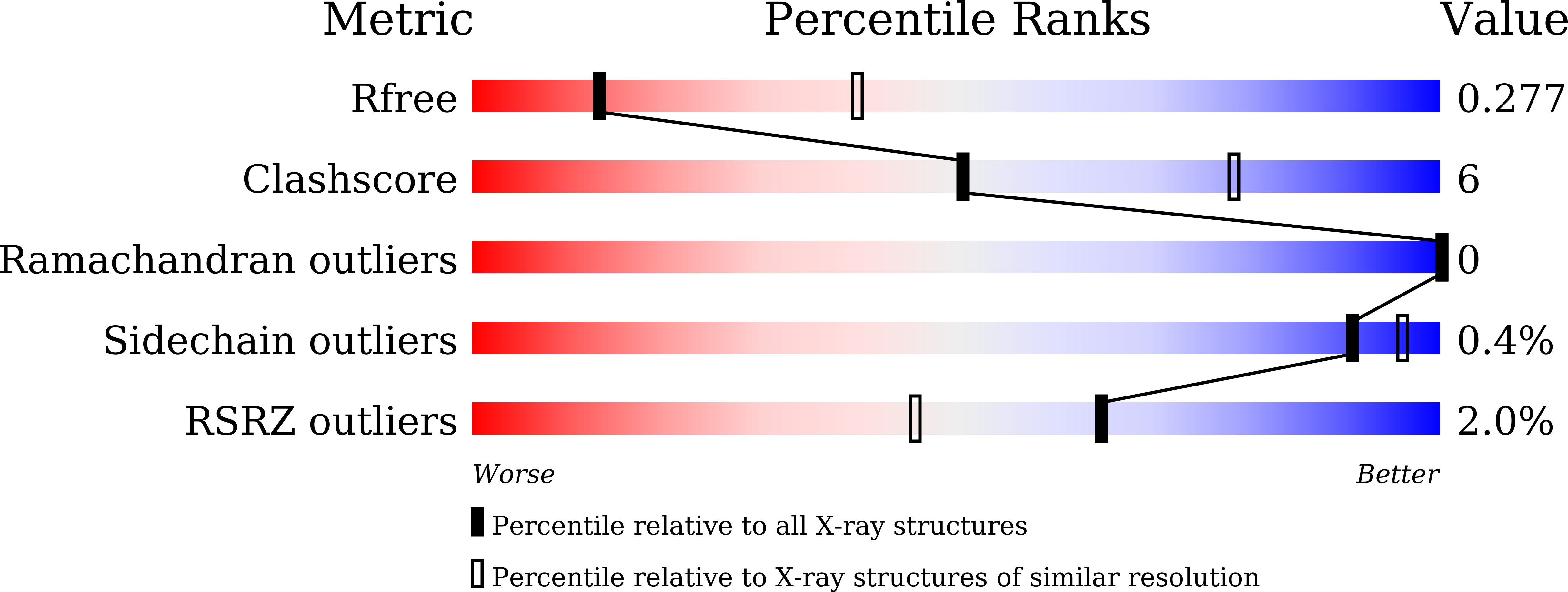
Resolution:
3.13 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.23
Space Group:
P 63 2 2