Deposition Date
2023-03-08
Release Date
2023-07-12
Last Version Date
2024-05-22
Entry Detail
PDB ID:
8GFK
Keywords:
Title:
Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
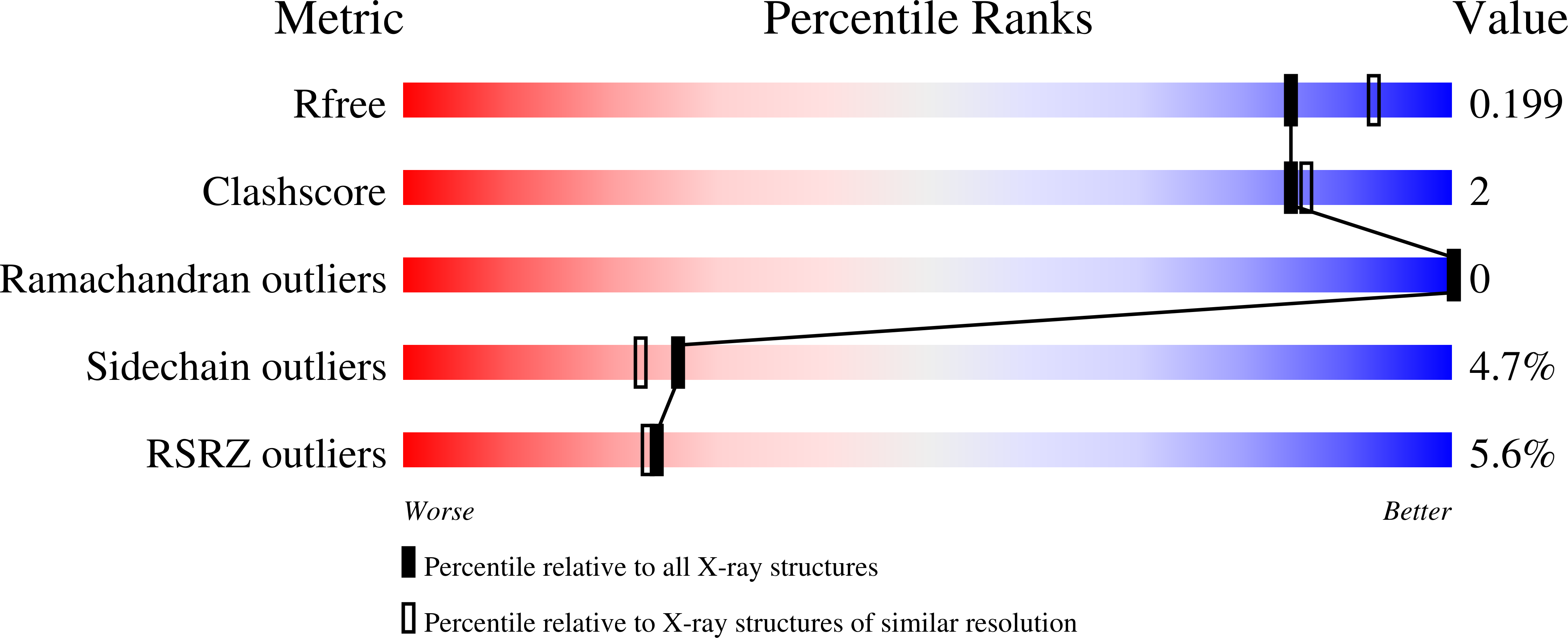
Resolution:
2.00 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.17
Space Group:
I 1 2 1