Deposition Date
2023-02-22
Release Date
2024-03-06
Last Version Date
2025-05-28
Entry Detail
PDB ID:
8G9S
Keywords:
Title:
Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications
Biological Source:
Source Organism(s):
Neisseria lactamica (Taxon ID: 486)
Rhodobacter phage RcNL1 (Taxon ID: 754047)
Rhodobacter phage RcNL1 (Taxon ID: 754047)
Expression System(s):
Method Details:
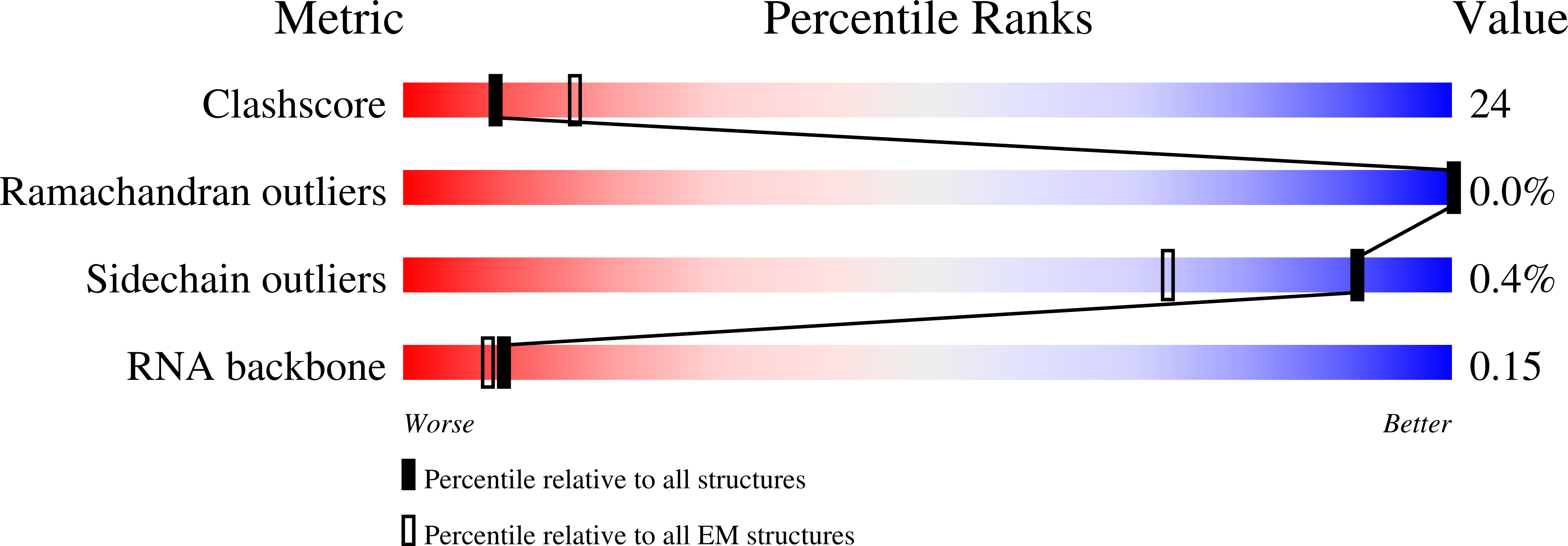
Experimental Method:
Resolution:
3.40 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE