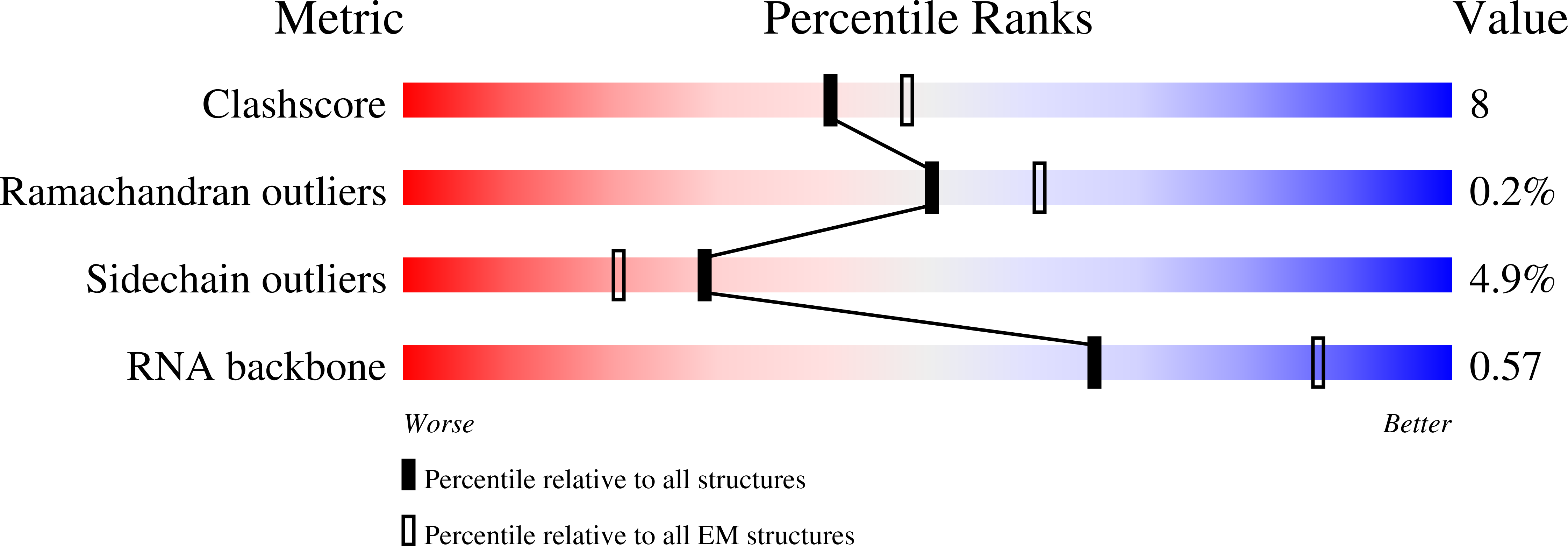
Deposition Date
2023-02-16
Release Date
2023-03-22
Last Version Date
2023-11-15
Entry Detail
PDB ID:
8G6X
Keywords:
Title:
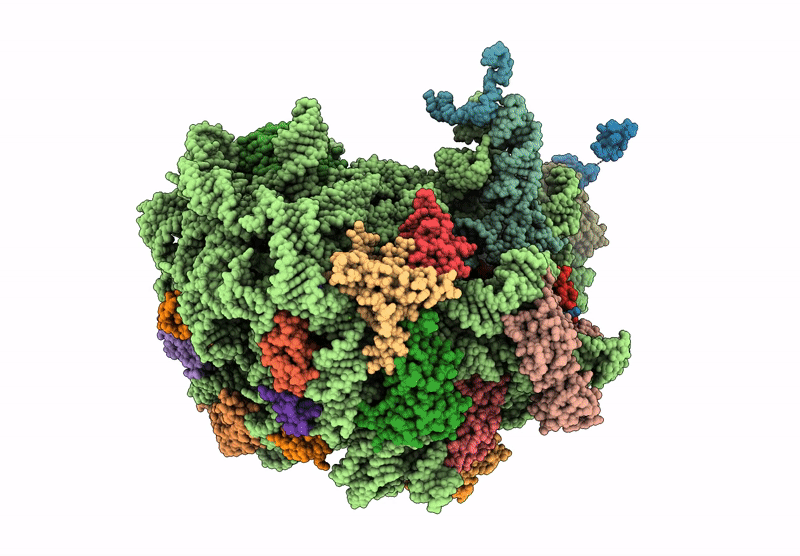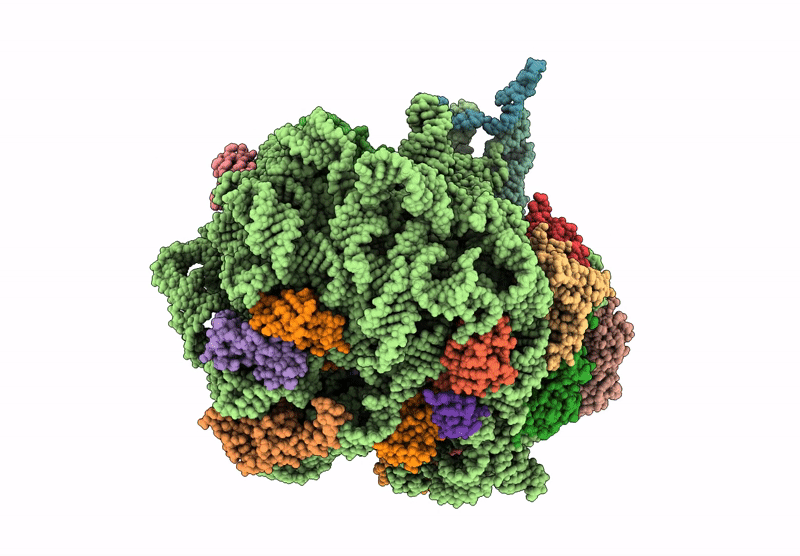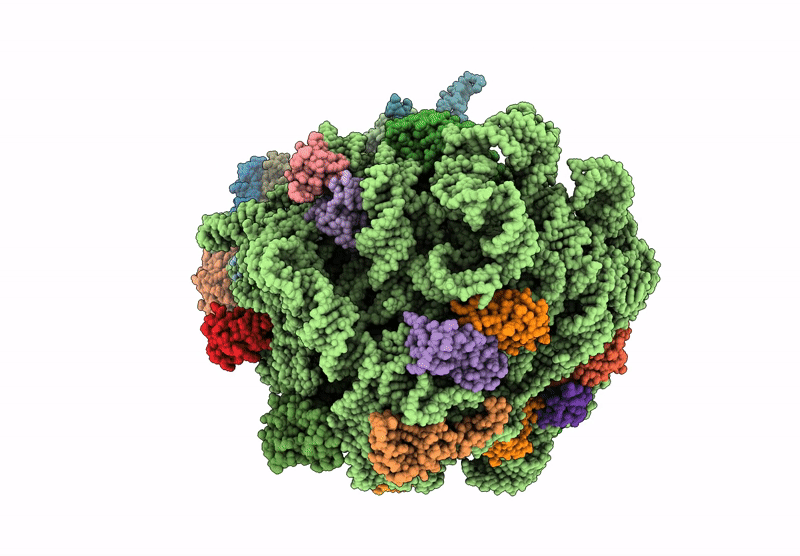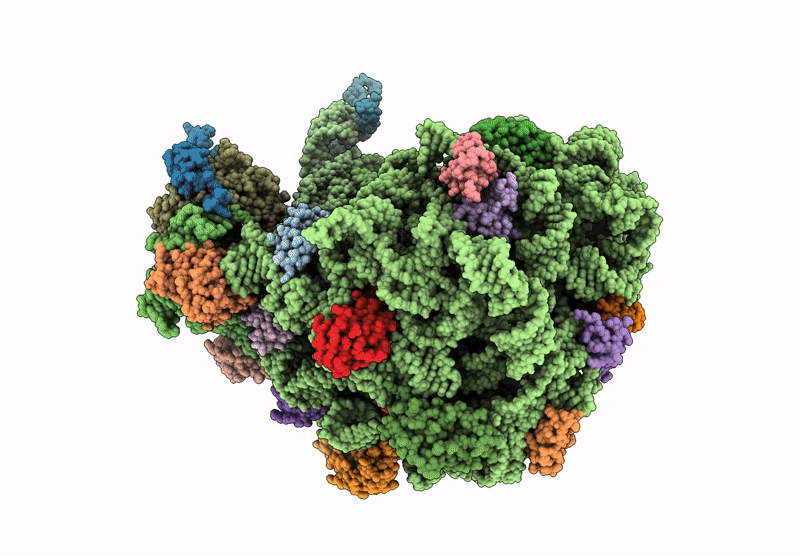
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site meta-aminobenzoic acid charged NH-tRNAPhe
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details:
Experimental Method:
Resolution:
2.31 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE